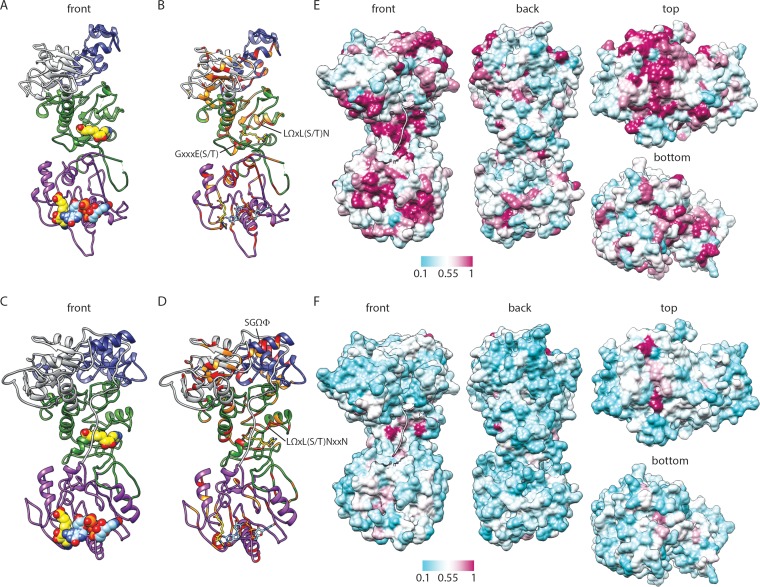
FIG 4.
Predicted structure and conservation for RRV VP3. (A to D) Ribbon drawings of front views of the RRV VP3 PHYRE homology (A and B) and I-TASSER threading (C and D) models, based on BTV10 VP4 (PDB accession number 2JH8). Predicted domains are colored as described in the legend to Fig. 2. BTV10 VP4 ligands SAH (yellow; PDB accession number 2JHP) and GpppG (light blue; PDB accession number 2JHA) have been overlaid as sphere (A and C) or stick (B and D) representations. Residues that are identical (red) or similar (orange) in structure-based alignments with BTV10 VP4 are indicated in panels B and D. Motifs identified in Fig. 2 or discussed in the text are indicated. (E and F) Four views of a surface representation of the VP3 I-TASSER threading model colored by the degree of amino acid conservation across clade A rotavirus (E) or all (F) sequences from the alignment in Fig. 2. The most highly conserved positions are colored maroon, and the least conserved are colored cyan, as indicated by the scale bar. The first nine amino acids of the model are shown as a ribbon.