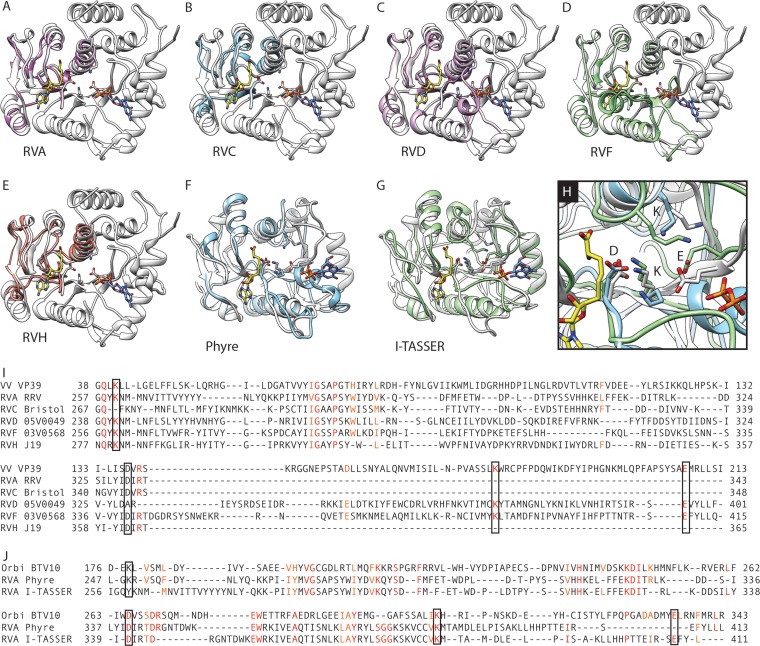
FIG 5.
Predicted 2′-O-methyltransferase domain of RRV VP3. (A to E) Overlay of ribbon drawings of VV VP39 (white; PDB accession number 1V39) and the VP39-based VP3 homology models for RVA RRV (A), RVC Bristol (B), RVD 05V0049 (C), RVF 03V0568 (D), or RVH J19 (E). The VP39 ligands SAH (yellow) and 7N-methyl-8-hydroguanosine-5′-diphosphate (light blue) are shown as stick representations. (F to H) Overlay of the 2′-O-MTase domain of the BTV10 VP4 homology model (white) with the VP4-based RVA RRV VP3 homology model (light blue; F), I-TASSER RVA RRV VP3 threading model (green; G), or both (H). The VP4 ligands SAH (yellow; PDB accession number 2JHP) and 7N-methyl-8-hydroguanosine-5′-diphosphate (blue; PDB accession number 2JH8) are shown. Side chains of residues in the KDKE motif are shown (A to H) and indicated (H). (I to J) Structure-based alignment of sequences from the VP3 models shown in panels A to E with VV VP39 (I) or from the RRV VP3 models shown in panels F and G with BTV10 VP4 (J). Identical (red) and similar (orange) residues are colored. Residues of the KDKE motif are outlined.