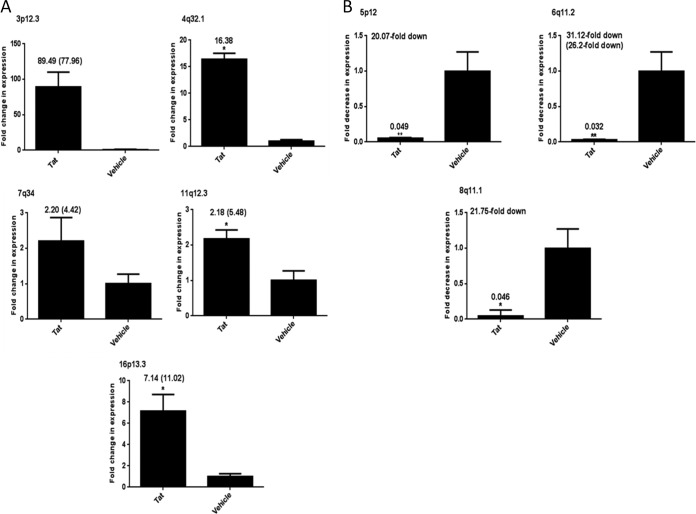
FIG 2.
Validation of RNA-Seq data for HERV-K(HML-2)-specific proviruses and HLA-A. Total cellular RNA was isolated from primary cells (PBLs) that were treated with recombinant Tat protein or vehicle alone for 12 h. RNA was DNase treated, and cDNA was synthesized and amplified by SYBR green two-step qRT-PCR. Data are expressed as fold increase in RNA (Tat treatment over vehicle alone) (A) or fold decrease in RNA (Tat treatment over vehicle alone) (B), with the fold written on top of the bar. Numbers in parentheses indicate the fold activation as measured by RNA-Seq for ease of comparison for those proviruses whose RPKM was not zero in either treatment. (C) Expression of HLA-A RNA in PBLs treated with Tat or vehicle using two groups of PBLs (3 replicates each). PBLs 1 to 3 had a 1.5-fold decrease in RNA expression (2.16-fold decreased by RNA-Seq), and PBLs 4 to 6 had a 0.28-fold increase in HLA-A RNA expression. All qRT-PCR results were normalized to those for the GAPDH reference gene after analysis using the 2−ΔΔCT method, and relative expression is plotted. Error bars indicate standard errors of the means for results from the PBLs of seven individual volunteers (except for panel C, which represents three individual samples each). Significance was calculated by comparing Tat treatment to vehicle treatment using a Student's t test. Significant results are indicated (*, P < 0.05).