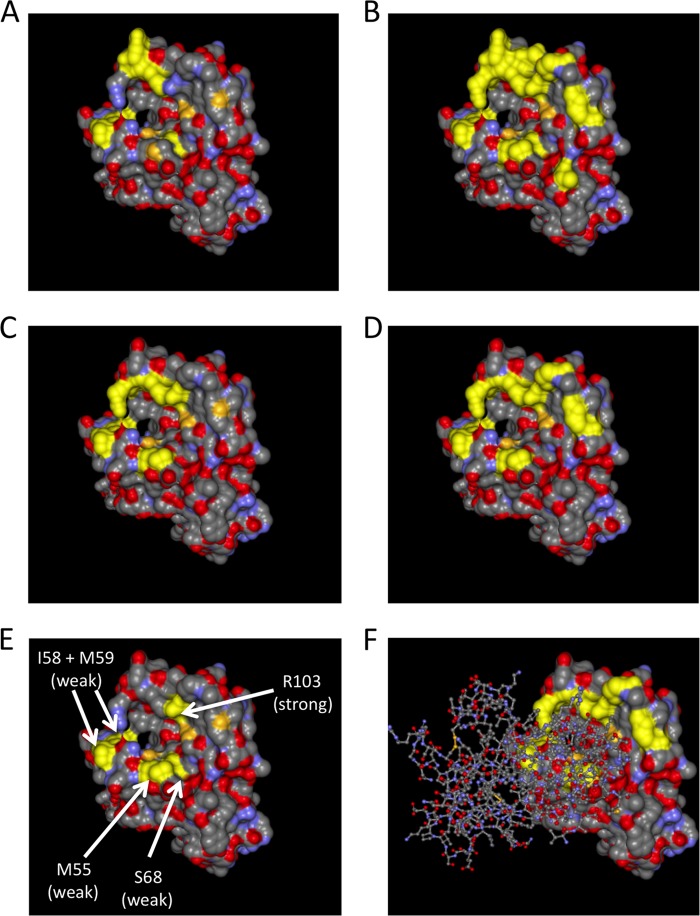
FIG 6.
Structure of the extracellular domain of EphA2 viewed from the top. The crystal structure of the EphA2 extracellular domain is represented as a solvent-excluded surface. (A) Residues I58, M59, S68, F156, and E157 on EphA2 are highlighted in yellow. These residues correspond to the binding interface of compound 1 on EphA4 as determined by Qin et al. (B) All residues that were mutated for binding experiments are highlighted in yellow. (C) All residues that exhibited a clearly visible decrease in binding to ephrin A4 upon mutation are highlighted in yellow. (D) All residues that exhibited a clear decrease in binding to ephrin A5 upon mutation are highlighted in yellow. (E) M55, the amino acid pair I58 and M59, S68, and R103, whose mutation affected binding of KSHV gH/gL, are highlighted in yellow. (F) Same representation as in panel D, but with a “stick and ball” model of the structure of the bound receptor binding domain of ephrin A5 superimposed.