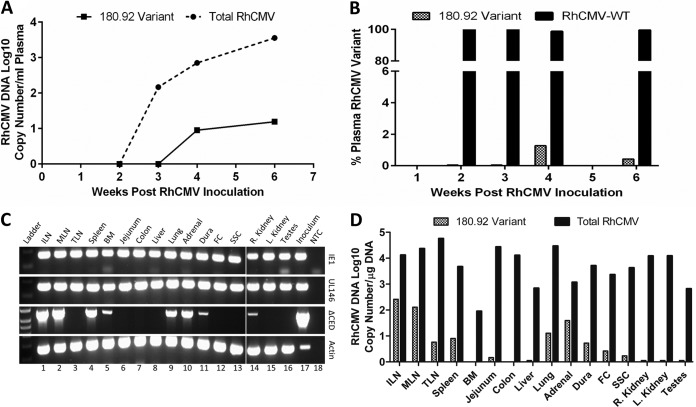
FIG 4.
Viral load and tissue distribution of RhCMV in a SIV-infected CMV-seronegative rhesus macaque experimentally inoculated with RhCMV 180.92. Real-time and conventional PCR data shown. (A) Median DNA copy numbers of total RhCMV plasma virus burden (solid line) and RhCMV 180.92 variant (dashed line). (B) Percentage of RhCMV 180.92 variant copy number (hashed column) compared to percentage of WT-like RhCMV variant (RhCMV-WT) copy numbers (solid column), as measured by variant-specific real-time PCR at 2, 3, 4, and 6 weeks after RhCMV 180.92 experimental inoculation. (C) Genomic DNA extracted from various tissues was assayed to differentiate total RhCMV (IE1), RhCMV-WT variant (UL146), and RhCMV 180.92 variant (ΔCED) using a variant-specific conventional PCR assay. Lane 18 is nontemplate control (water [NTC]) serving as a negative control, and DNA extracted from RhCMV 180.92 virus stock (Inoculum, lane 17) and β-actin amplifications (Actin) served as positive controls. (D) Quantitative analysis of RhCMV 180.92 variant DNA copy number/μg of DNA as measured by real-time PCR of 100 ng of tissue genomic DNA. Median values determined for three replicate wells are shown. ILN, inguinal lymph node; MLN, mesenteric lymph node; TLN, tracheobronchial lymph node; BM, bone marrow; FC, frontal cortex; SSC, sacral spinal cord.