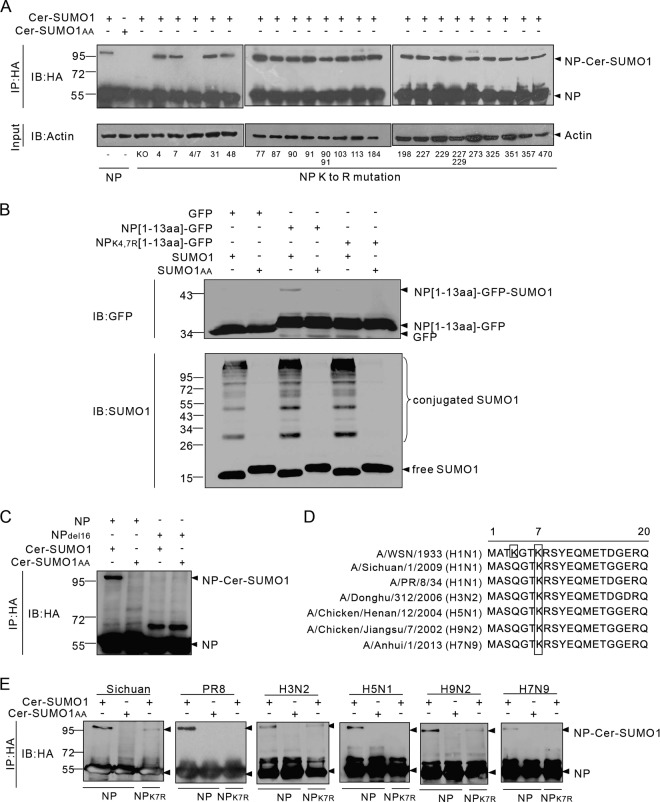
FIG 2.
K4 and K7 are SUMO acceptor sites for different influenza A virus subtypes and strains. (A) Expression plasmids encoding WSN-WT NP or NP with K-to-R mutations at the indicated positions and Ubc9 were transfected together with Cer-SUMO1 or Cer-SUMO1AA. The cells were lysed, and NP was immunoprecipitated (IP) with anti-HA–agarose and then detected by Western immunoblotting (IB) with anti-HA polyclonal antibodies. (B) The expression plasmid pCAGGS-GFP, NP[1-13aa]-GFP, or NPK4,7R[1-13aa]-GFP was cotransfected with SUMO1 or SUMO1AA; Ubc9 was also cotransfected with each pCAGGS plasmid. The cells were lysed and directly subjected to Western blotting with anti-GFP or anti-SUMO1 antibodies. (C) Expression plasmids encoding WSN-WT NP or the NPdel16 mutant form and Ubc9 were transfected together with Cer-SUMO1 or Cer-SUMO1AA. The cells were lysed, and NP was immunoprecipitated with anti-HA–agarose and then detected by Western immunoblotting with anti-HA polyclonal antibodies. (D) Schematic representation of the N-terminal amino acid sequences of NPs from different influenza A virus strains. The lysine residues (framed) indicate the SUMO1 modification sites for each NP. (E) 293T cells were cotransfected with plasmids encoding Ubc9 and HA-tagged NPs or the NPK7R mutant form from different influenza A virus subtypes and strains together with Cer-SUMO1 or Cer-SUMO1AA. The cells were lysed, and NP was immunoprecipitated with anti-HA–agarose. The precipitated proteins were further analyzed by SDS-PAGE and Western blotting with anti-HA polyclonal antibodies. The values to the left of the blots are molecular sizes in kilodaltons.