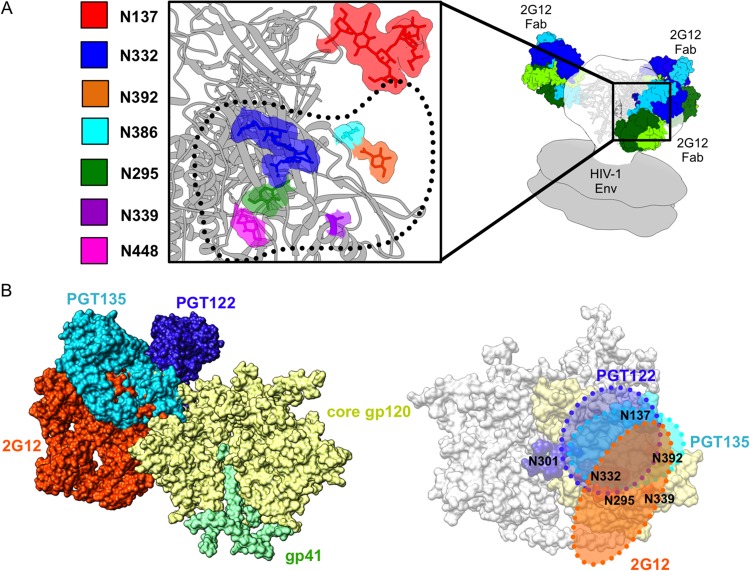
FIG 3.
Details of the glycan epitope of 2G12. (A) A closeup view of the interface between BG505 SOSIP.664 Env (PDB ID 4NCO) and 2G12 Fab2 (PDB ID 1OP5) (contact interface highlighted with dotted black line) showing the 2G12-relevant glycans. Glycans from the Env crystal structure that fall within the 2G12 epitope are rendered in space-filling mode and highlighted by different colors as indicated in the key. gp120 is rendered in gray ribbon. Our 2G12-Env EM reconstruction was fit into a model of unliganded membrane-anchored HIV-1 trimer (gray surface) (EMDB accession numbers EMD-5019 and EMD-5021) (60) in order to fit the Fab2 crystal structures. The BG505 SOSIP.664 crystal structure was subsequently fit into our EM density map in order to determine which glycans fell within the 2G12 epitope (20). The crystal structures used in the docking were solved after partial deglycosylation, such that some of the N-linked glycans only harbor an N-acetylglucosamine (NAG), while others contain high-mannose glycans (20). (B) On the left, we show a superimposition of glycan-dependent monoclonal antibody (MAb) cocrystal structures of PGT135 (PDB ID 4JM2) (13) and PGT122 (PDB ID 4NCO) fit onto the Env trimer crystal structure along with our own EM fitting of 2G12 to expand on the mapping of this glycan supersite of vulnerability on Env, showing the overlap of these three antibodies. The extents of these epitopes are detailed on the right, further emphasizing the large size of the 2G12 epitope on the trimeric surface of Env and where it overlaps with other glycan-dependent MAbs.