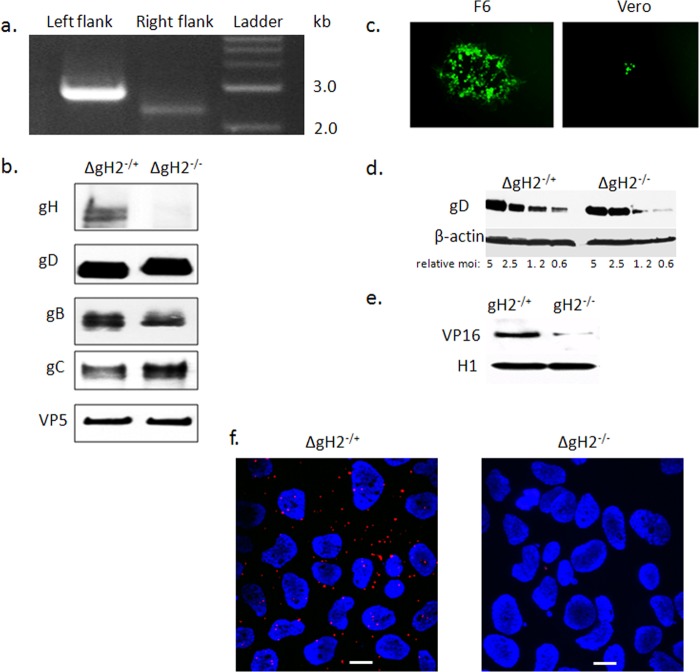
FIG 5.
Construction and characterization of an HSV-2 gH-null virus. (a) Genotypic characterization of ΔgH-2 was performed by PCR using two primer sets to confirm appropriate replacement of gH. The left flank of UL22 was tested with primers gH2-L-check and sacB-Out, while the right flank of this gene was assessed with primers gH2-R-check and Hyg-Out (Table 1). (b) The gH-null virus was propagated on gH-1-expressing F6 cells to yield complemented virus (ΔgH−/+) or on Vero cells to yield noncomplemented virus (ΔgH−/−). The viruses were purified on sucrose gradients, and equivalent numbers of viral particles (estimated by comparing VP5 expression on Western blots) were analyzed for expression of viral proteins by immunoblotting with rabbit polyclonal Ab to gH-gL or murine MAbs to gB, gD, gC, and VP5. (c) Representative fluorescence microscopy image obtained 36 h p.i. of F6 or Vero cells with ΔgH−/+. (d) To evaluate whether deletion of gH impacted binding, CaSki cells were exposed to serial 2-fold dilutions of relatively equal numbers of purified complemented or noncomplemented gH-null viruses (starting with viral particle numbers equivalent to an MOI of 5 PFU/cell) at 4°C for 4 h. Binding was assessed by performing Western blots of cellular lysates with gD and β-actin, and results shown are representative of 3 independent experiments. (e) CaSki cells were inoculated with purified virus (relative particle numbers equivalent to an MOI of 1 PFU/cell on F6 cells), and nuclear extracts were prepared 1 h p.i. and probed for the tegument protein VP16 and histone 1 (H1). A blot representative of results from 3 independent experiments is shown. (f) CaSki cells were synchronously infected with purified ΔgH−/+ or ΔgH−/− (equivalent to 5 PFU/cell) and fixed and probed with monoclonal mouse antibodies to integrin αvβ3 and rabbit sera to gH-gL and assessed in a proximity ligation assay. Results are representative of 3 independent experiments.