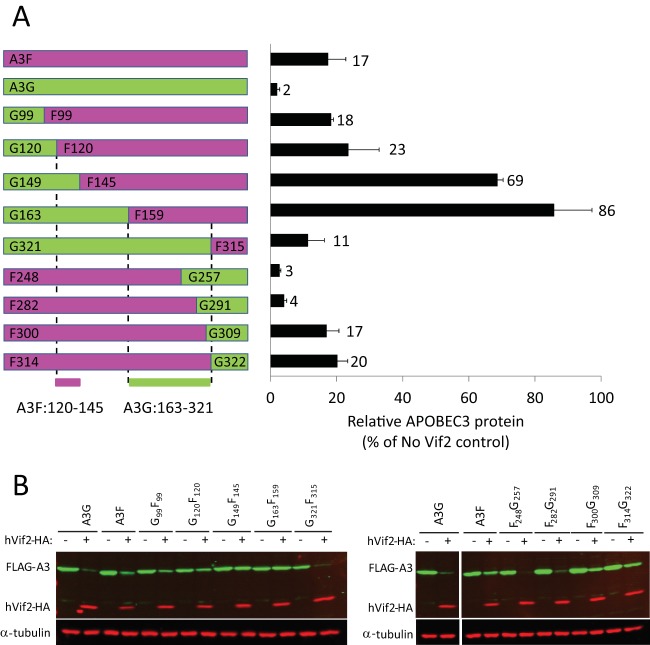
FIG 7.
Sensitivity of A3G/A3F and A3F/A3G chimeras to hVif2-HA. (A) Schematic structures of A3G/A3F (GF) and A3F/A3G (FG) chimeras. Full-length A3G and the A3G portions of the GF and FG chimeras are shown in green, whereas the full-length A3F and A3F portions of the chimeras are shown in fuchsia. The numbers indicate the amino acid position from each A3 protein at which the A3G and A3F portions were fused to generate the chimeras; for example, the G99F99 chimera contains amino acids 1 to 99 from A3G and amino acids 99 to 373 from A3F. Quantitation of hVif2-HA-induced degradation of each chimera is shown in the bar graph to the right of the schematic structures of the chimeras. The relative amount of each A3 protein remaining in the presence of absence of hVif2-HA was determined by quantitative Western blotting, and the amount of each A3 protein in the absence of Vif2 was set equal to 100%. Data are plotted as the average of two independent experiments, and error bars indicate standard deviations. (B) Representative images of Western blots indicating sensitivity to Vif2-HA are shown. Cell lysates were analyzed by using an anti-FLAG antibody to detect FLAG-A3 or an anti-α-tubulin antibody to detect α-tubulin. Lanes +, with Vif2; lanes −, without Vif2. Comparison of the Vif2 sensitivity of the G120F120 and G149F145 chimeras indicates that the determinants in A3F that confer sensitivity to Vif2 are between amino acids 121 and 144; a comparison of the G163F159 and G321F315 chimeras indicates that the determinants in A3G that confer sensitivity to Vif2 are between amino acids 163 and 321.