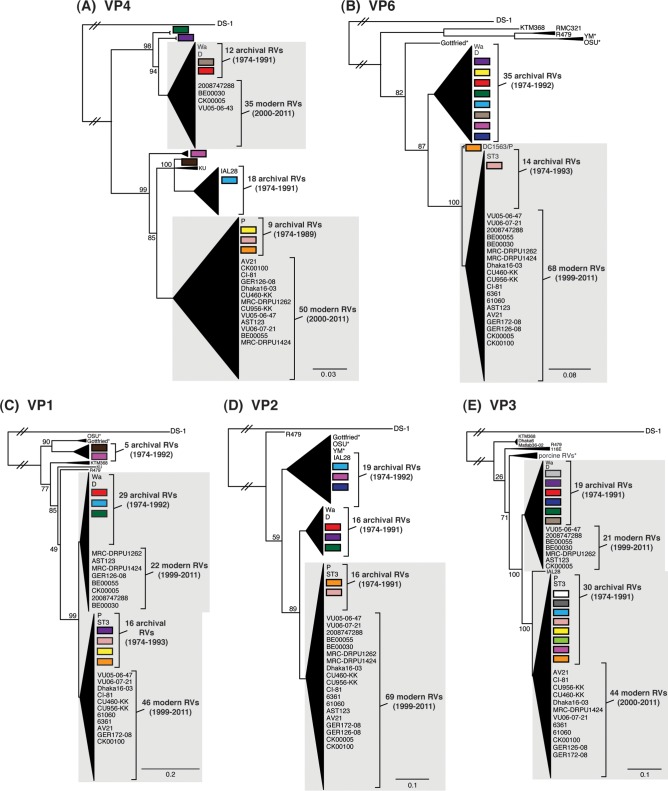
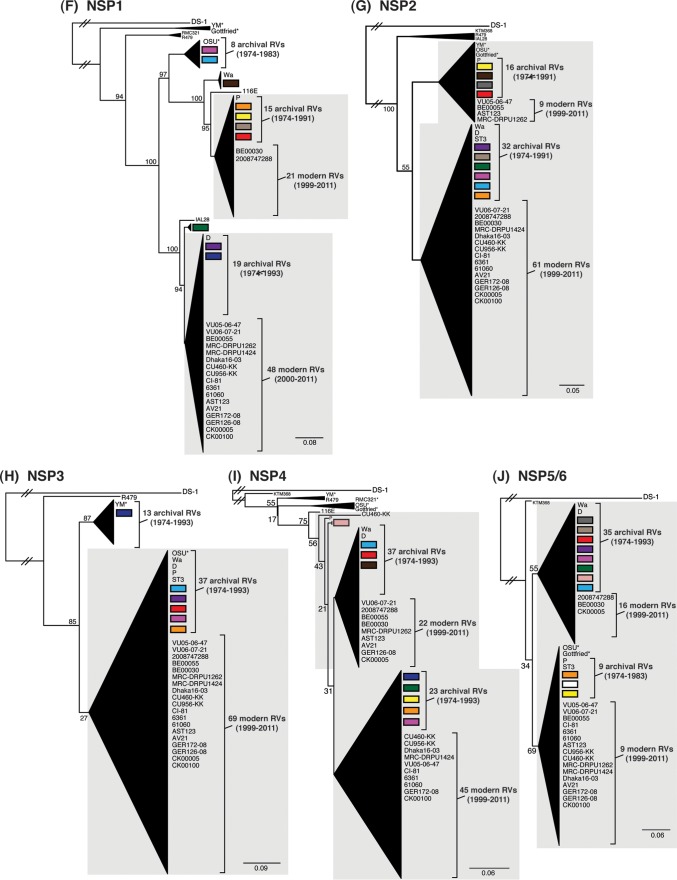
FIG 4.
Comparison of the VP1 to VP4, VP6, and NSP1 to NSP5/6 genes from the DC RVs to those of other strains. Maximum-likelihood intragenotypic phylogenetic trees were constructed for each gene and are outgroup rooted to the genes of strain DS-1. The following nucleotides were used for each gene: VP4 gene, nucleotides 10 to 2337 (A); VP6 gene, nucleotides 24 to 1217 (B); VP1 gene, nucleotides 19 to 3267 (C); VP2 gene, nucleotides 155 to 2701 (D); VP3 gene, nucleotides 50 to 2557 (E); NSP1 gene, nucleotides 32 to 1492 (F); NSP2 gene, nucleotides 47 to 1000 (G); NSP3 gene, nucleotides 35 to 967 (H); NSP4 gene, nucleotides 42 to 569 (I); and NSP5/6 gene, nucleotides 21 to 614 (J). Horizontal branch lengths are drawn to scale (nucleotide substitutions per base), bootstrap values are shown as percentages for key nodes, and monophyletic groupings were collapsed (triangles). The relative locations of the subgenotype alleles defined for the DC RVs in the trees are shown as colored rectangles. The relative locations of archival and modern RV genes in each tree are indicated, and select RV strains are listed. The asterisks indicate animal RV strains.