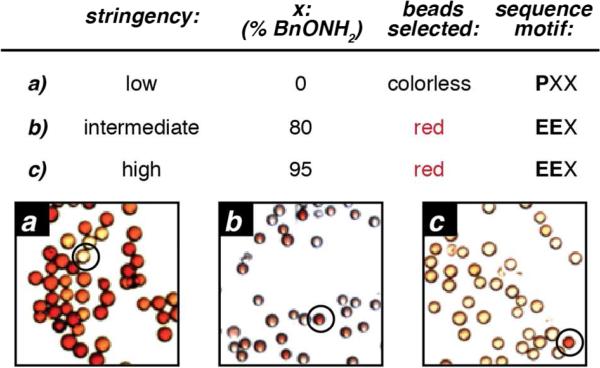
Figure 3.
Library screening to identify reactive sequences towards Rapoport's salt-mediated transamination. The library was treated with 10 mM RS for 1 h at pH 6.5, followed by oxime formation with the specified ratios of BnONH2 (x%) and DispRed-ONH2 (100–x%). The active sequences were identified by selecting and sequencing the red beads in the library. When a low stringency screen was applied (a) many beads had a red color, indicating that many sequences form some degree of oxime product. In this case, the beads that remained colorless were selected, and proline was identified as an N-terminal residue that prevented reactivity. To identify sequences that led to high levels of oxime product, more stringent screens were used that could distinguish low from high levels of oxime yield (b, c). Sequencing the red beads from these screens identified glutamate-terminal sequences as a common reactive motif.