Fig. 5.
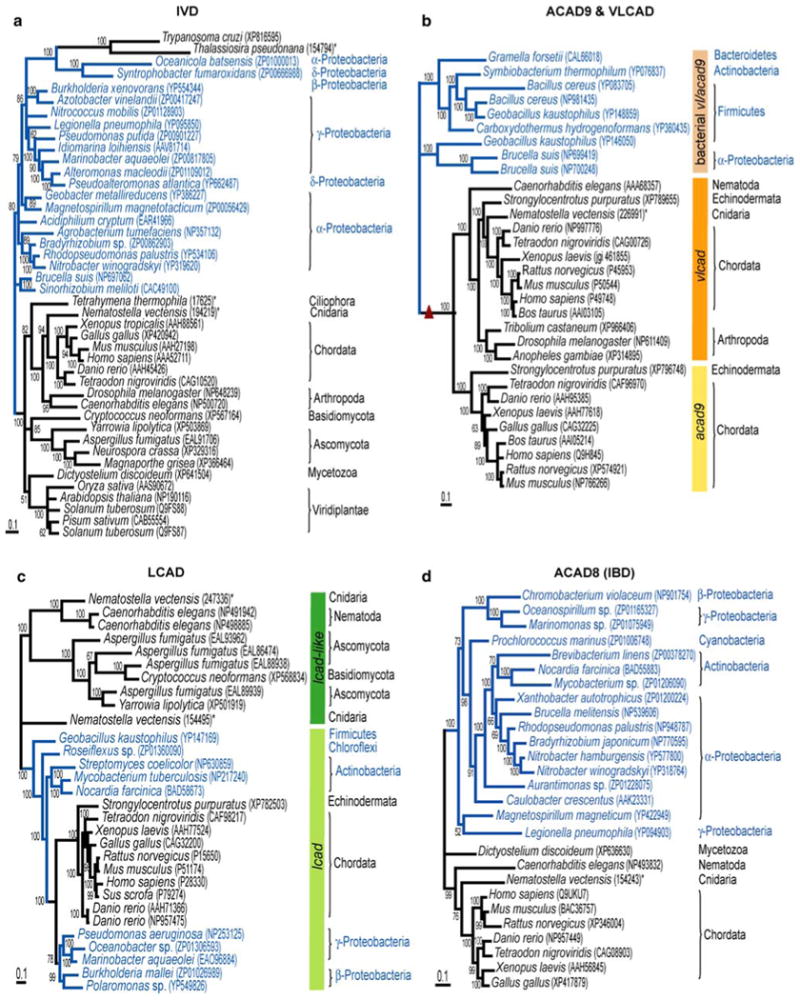
Bayesian phylogeny of isovaleryl (a), very long-chain and ACAD9 (b), long-chain (c), and isobutyryl-CoA (d) dehydrogenase subfamilies. Unrooted phylograms were estimated from 384, 544, 375, and 373 amino acids, respectively. Numbers at internodes show posterior probabilities for Bayesian consensus tree. Small triangle in b refers to gene duplication event
