Table 1.
The Fat-like cadherin CDH-4 controls AQR and PQR migration
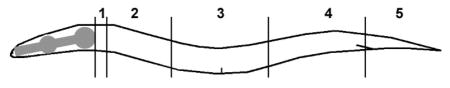
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Genotype | AQR position (%) | PQR position (%) | ||||||||||
|
|
|
|||||||||||
| 1 | 2 | 3 | 4 | 5 | N | 1 | 2 | 3 | 4 | 5 | N | |
|
|
|
|||||||||||
| wild-type | 100 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 100 | 100 |
| cdh-4(lq56) | 39 | 3 | 2 | 7 | 49 | 100 | 28 | 3 | 4 | 4 | 60 | 100 |
| cdh-4(lq56); Ex[cdh-4(+)] | 90* | 0 | 1 | 0 | 9 | 100 | 2* | 0 | 1 | 1 | 96 | 100 |
| cdh-4(lq83) | 48 | 2 | 0 | 3 | 47 | 100 | 23 | 0 | 0 | 2 | 75 | 100 |
| cdh-4(lq97) | 80** | 3 | 1 | 0 | 16 | 205 | 19 | 2 | 1 | 1 | 77 | 205 |
| cdh-4(lq97); smg-1(RNAi) | 82 | 3 | 1 | 0 | 14 | 200 | 13 | 1 | 1 | 1 | 84*** | 200 |
| cdh-4(rh310) | 50 | 1 | 1 | 2 | 46 | 100 | 19 | 0 | 0 | 2 | 79 | 100 |
| cdh-4(ok1323) | 45 | 0 | 2 | 1 | 52 | 100 | 24 | 0 | 1 | 0 | 75 | 100 |
| cdh-4(hd40) | 42 | 1 | 1 | 2 | 54 | 100 | 20 | 0 | 0 | 1 | 80 | 100 |
| cdh-4(RNAi) | 100 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 100 | 100 |
| unc-40(n324) | 96 | 1 | 0 | 0 | 3 | 76 | 32 | 3 | 8 | 6 | 51 | 76 |
|
unc-40(n324); cdh-4(RNAi) egl-20(n585) |
99 | 0 | 0 | 1 | 0 | 100 | 76 | 4 | 2 | 5 | 13**** | 100 |
| cwn-2(ok895); cwn-1(ok546) | 39 | 49 | 10 | 2 | 0 | 100 | 3 | 16 | 19 | 25 | 37 | 100 |
p < 0.0001 compared to other cdh-4(lq56).
p < 0.001 compared to other cdh-4 alleles lq56, lq83, rh310, ok1323, and hd40.
p = 0.0416 compared to cdh-4(lq97) alone.
p < 0.0001 compared to unc-40(n324) alone.
