Figure 2. Structural differences between tRNA and the TLS.
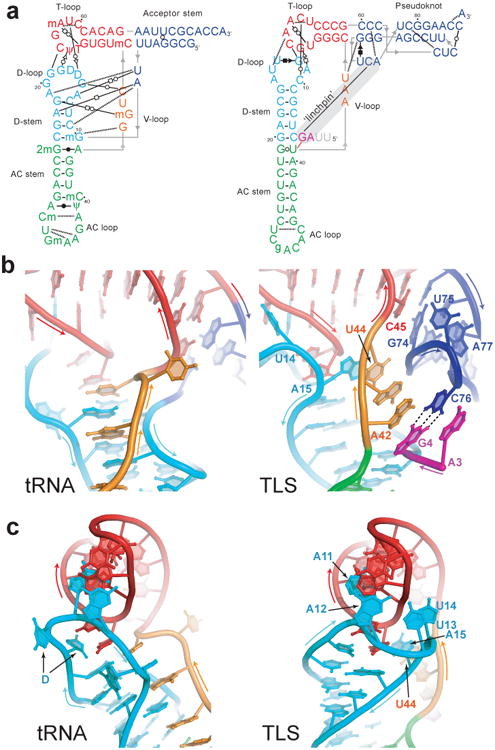
a, Secondary structures showing interactions that stabilize the folds of tRNA (left) and the TLS (right). Non-canonical base pairs are indicated with Leontis-Westhof symbols30, single hydrogen bonds with dashed lines. Lines with embedded arrows indicate chain connectivity. Grey nucleotides were not visible in the electron density. A grey bar indicates the long-range ‘linchpin’ interaction. b, Intramolecular interactions of the V-loop (orange) in tRNA (left) and the TLS (right). Dashed lines indicate the C76-G4 base pair. c, Conformation and interaction of the D-loop (cyan) with the T-loop (red) of tRNA (left) and the TLS (right).
