Extended Data Figure 6. T-loop and acceptor stems of the tRNA and TLS and elongation factor binding.
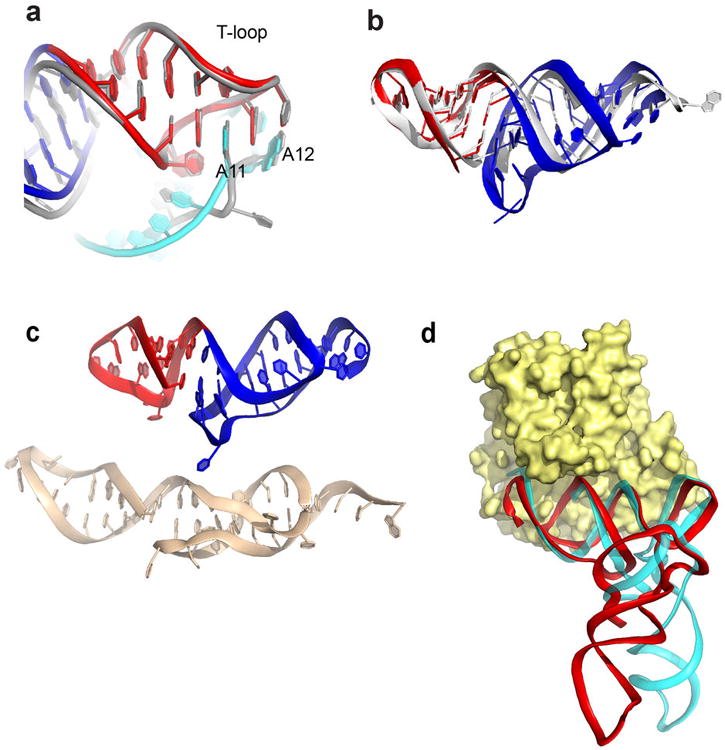
a, Superimposed structures of the TLS T-loop (red) and part of the D-loop (cyan) with the analogous structures in tRNA (grey). TLS bases A11 and A12 are shown; these bases match the interactions formed by analogous bases in tRNA. In the TLS, A11 is in a syn conformation, but the matching base in tRNA is not. This may be due to local differences in the backbone conformation. b, Superimposed structures of the TLS T-loop (red) and pseudoknot (blue) with the T-loop and acceptor stem elements in a tRNA (grey). View is from the “top” of the molecule, down the axis of the D- and AC-stems. c, Top: The structure of the T-loop (red) and acceptor stem pseudoknot (blue) in the TLS crystal structure. Bottom: structure of these elements isolated from the rest of the TLS and solved by NMR (PDB: 1A60)43. d, Superposition of the TLS structure (red) onto the tRNA (cyan) of a tRNAPhe bound to EF-Tu (yellow), the bacterial homolog of eEF1A (PDB: 1TTT)44. Binding is likely facilitated by the fact that RNA backbone conformation of the TLS pseudoknot and T-stem/loop matches that of a tRNA.
