Extended Data Figure 9. Models of protein binding to the TLS and the location of the UPD.
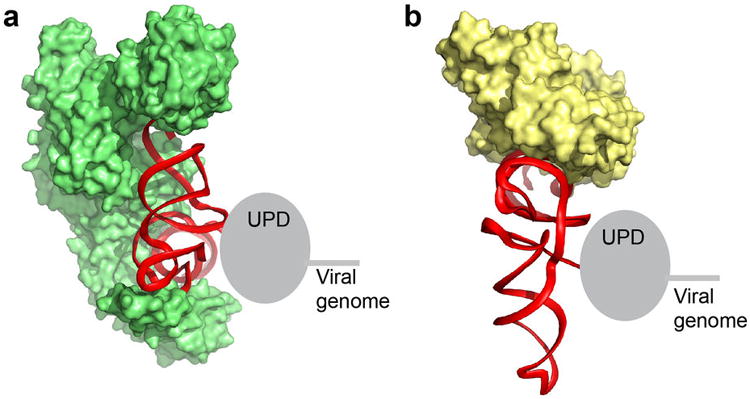
a, Model of the TLS (red, backbone ribbon shown) on the valine AARS (green; PDB: 1GAX), similar to Figure 3b, but viewed from the top and with the tRNAVal not shown. The location of the UPD directly 5′ of and against the TLS is shown as a grey oval. The viral genomic RNA is 5′ of the UPD. Note that the strategy used by the TYMV TLS to interact with this protein is likely very different from that used by the TLSs that are histidylated or tyrosylated, which are very different in terms of their secondary structure and fold6,9. b, Same as panel a, but with the TLS modeled onto the bacterial homolog of eEF1A (EF-Tu) as in Extended Data Fig. 6. tRNAPhe is not shown. In both complexes, the location of the 5′ end, the UPD, and viral genome would not interfere with protein binding. This would not be true if the TLS had a tRNA-like topology with the 5′ end paired to the 3′ end.
