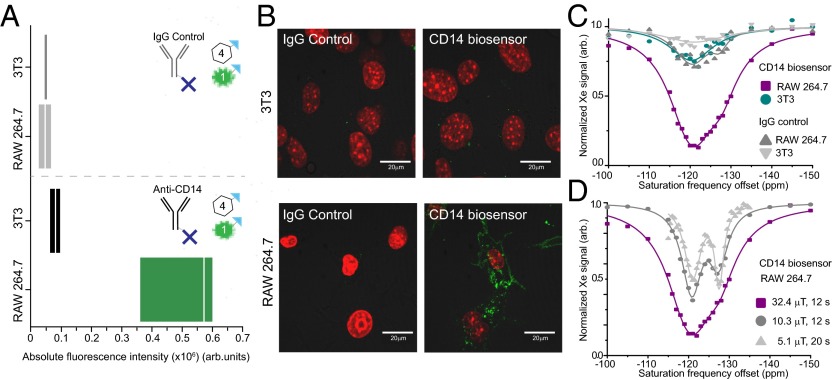
Fig. 2.
Dual readout modules facilitate evaluation of specificity and localization of the CD14 biosensor. Cells are sequentially incubated with the biosensor modules. First, cells are incubated with the targeting module, anti-CD14 antibody, or control, IgG antibody, both of which have been previously conjugated to avidin molecules (one avidin molecule per antibody depicted for simplicity). Secondly, cells are incubated with the readout modules in a 4:1 ratio of CrA−biotin:fluorescein−biotin. (A) The absolute fluorescence of the cell populations was evaluated by flow cytometry via the fluorescein−biotin readout module and shows the biosensor has good specificity for the target RAW 264.7 cell line. Median indicated by white line; range denotes first and third quartile. (B) Localization of the biosensor bound to cells was detected via fluorescence microscopy of the fluorescein−biotin readout module (green). Cell nuclei stained with Hoechst (red). (C) Xenon CEST spectroscopy (using a saturation pulse of 32.4 μT for 12 s) of cells shows a specific response at the resonance frequency for CrA, confirming the specific binding of the CrA−biotin module to the target RAW 264.7 cell line. (D) Using low-power saturation pulses (of 32.4 μT for 12 s, 10.3 μT for 12 s, and 5.1 μT for 20 s, respectively) we can reveal two peaks that correspond to CrA within a lipidic/cellular environment (left peak) and CrA within an aqueous environment (right peak).