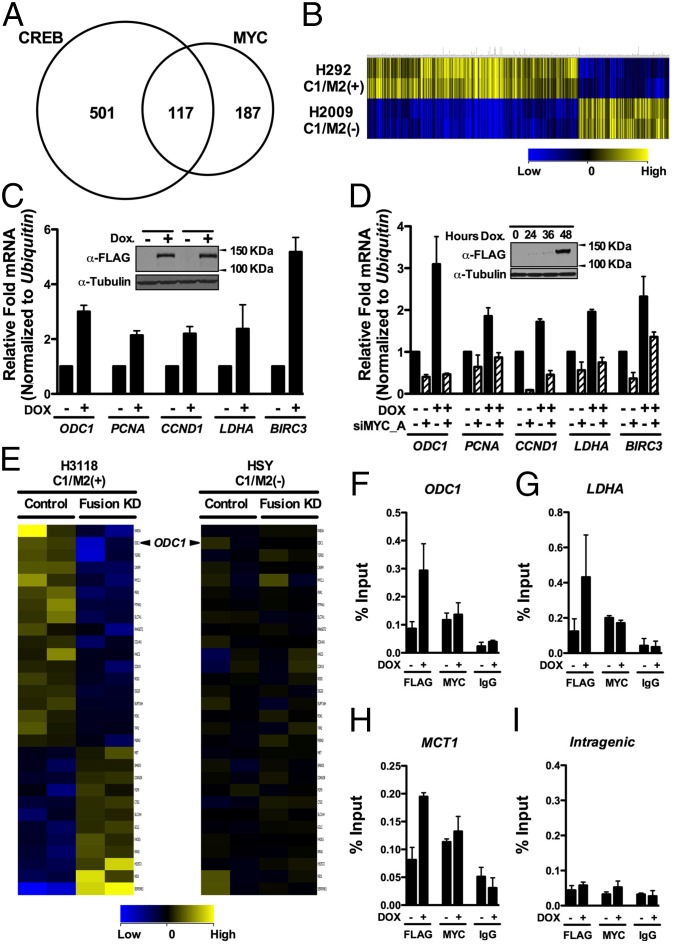
Fig. 2.
MYC target genes are regulated by the C1/M2 oncoprotein. (A) RNA-seq analysis of genes regulated by a stably integrated, Dox-inducible C1/M2 transgene in FLP-In T-Rex HEK293 cells (HEK293-CMVTetRTetOC1/M2) reveals that C1/M2 regulates genes of the CRTC:CREB and MYC:MAX networks. A Venn diagram is shown, with values representing the number of differentially regulated genes that are direct CREB and/or direct MYC targets. In total, 187 genes induced by C1/M2 were scored as direct MYC targets. (B) Hierarchical clustering of differentially expressed C1/M2-regulated CREB–MYC signature genes in H292 MEC (C1/M2+) versus H2009 non-MEC tumor cells. guanine cytosine robust multi-array analysis (GCRMA) quantile normalization was applied to the raw CEL files for two biological replicates from H2009 and H292 MEC cell line samples with baseline transformation set to the median of all samples. (C) Real-time qPCR analysis of endogenous MYC target genes in HEK293-CMVTetRTetOC1/M2 cells +/− Dox. Fold induction is shown; expression was normalized to Ubiquitin mRNA levels (n = 4). (Inset) Western blot analysis of C1/M2 levels following Dox treatment in independently derived stable cell clones. (D) Knockdown of MYC (siMYC_A) blocks the induction of Myc target genes by C1/M2 in double stable TRE-Tight-C1/M2 Tet-On Advanced A549 NSCLC lung cancer cells (A549-CMVrtTA2-M2TRE-TightC1/M2 cells). These cells were transfected with siMYC_A or nonspecific siRNAs (NSsi; −siMYC lanes) and then were treated +/− Dox for 48 h, and expression of MYC targets was assessed by real-time qPCR. The fold induction relative to nonspecific silencing RNA (NSsi) no Dox treatment is shown, and data were normalized to Ubiquitin mRNA levels (n = 4). (Inset) Western blot analysis of C1/M2 levels following Dox treatment. (E) Gene expression profiling of the C1/M2-regulated MYC signature genes in H3118 MEC tumor cells that harbor the t (11, 19) translocation and express C1/M2 compared with HSY tumor cells that lack the translocation and do not express C1/M2 following lentiviral-mediated delivery of C1/M2 (Fusion KD) or control shRNAs (n = 2 biological replicates). (F–I) C1/M2 is recruited to endogenous MYC-responsive promoters. Chromatin from HEK293-CMVTetRTetOC1/M2 cells +/− Dox (48 h) was immunoprecipitated with α-FLAG or α-MYC antisera, or with isotype-matched normal IgG, and analyzed for occupancy of the promoter-regulatory regions of the (F) ODC1, (G) LDHA, and (H) MCT1 genes relative to occupancy of a nonspecific (I) intragenic region of CCNB1 (84). Real-time qPCR quantification of C1/M2 and MYC occupancy is expressed as percent chromatin precipitated relative to input (n = 4). Data in C and D as well as F–I represent mean ± SEM.