Figure 1. A. SWCNT interactome.
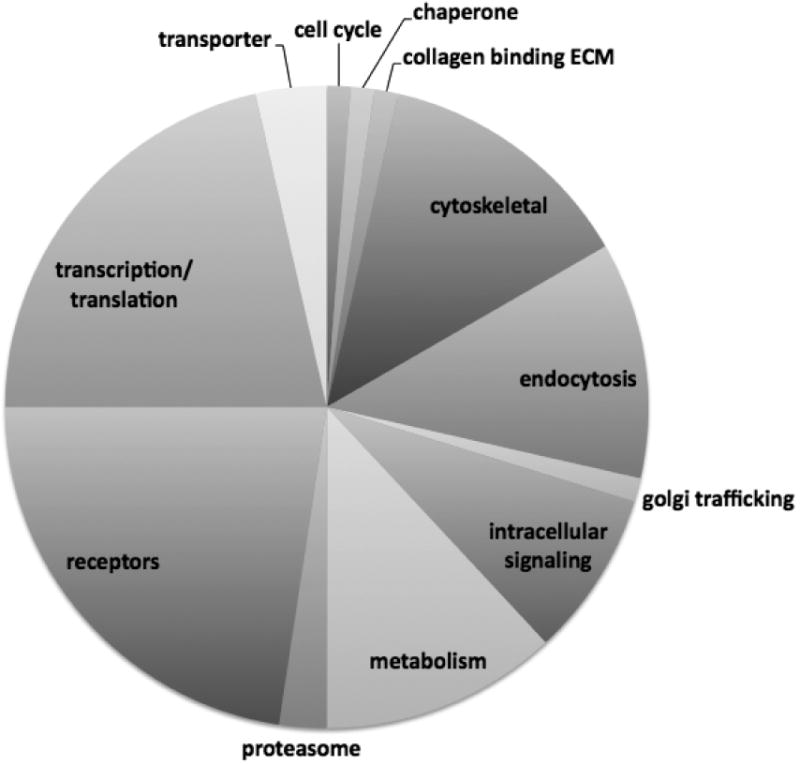

Ten micrograms of SWCNT were used as an affinity matrix to affinity purify, in triplicate, RBL2H3 cell lysates. An irrelevant matrix was used as a negative control. Interacting protein sets (approximately 27,000 peptides) were identified using complex mixture analysis and MuDPiT (tryptase digest and LCMS/MS) at the Keck Biotechnology Facility, Yale University. Canonical groups were identified using Panther and protein groups that were over represented in the SWCNT rather than control mixtures are shown as percentage distribution within canonical groups (pie chart). Exemplar proteins for some of the groups are shown in boxes, and numbers in parentheses represent multiple occurrences of a particular protein class or type within the set of peptides over-represented in CNT affinity purifications relative to controls. B. Chemical characteristics of previously characterized CNT binding peptides. Peptides shown to bind directly to CNT were collated from the literature (Wang et al., 2003, Jagota et al., 2002, 2006) and each residue from N- to C-termini was given a position number and color coded as follows: hydrophobic-aliphatic ●, hydrophobic-aromatic ●, polar neutral ●, hydroxyl or sulfur containing ●, acidic ●, basic ● and unique – cyclic ●.
