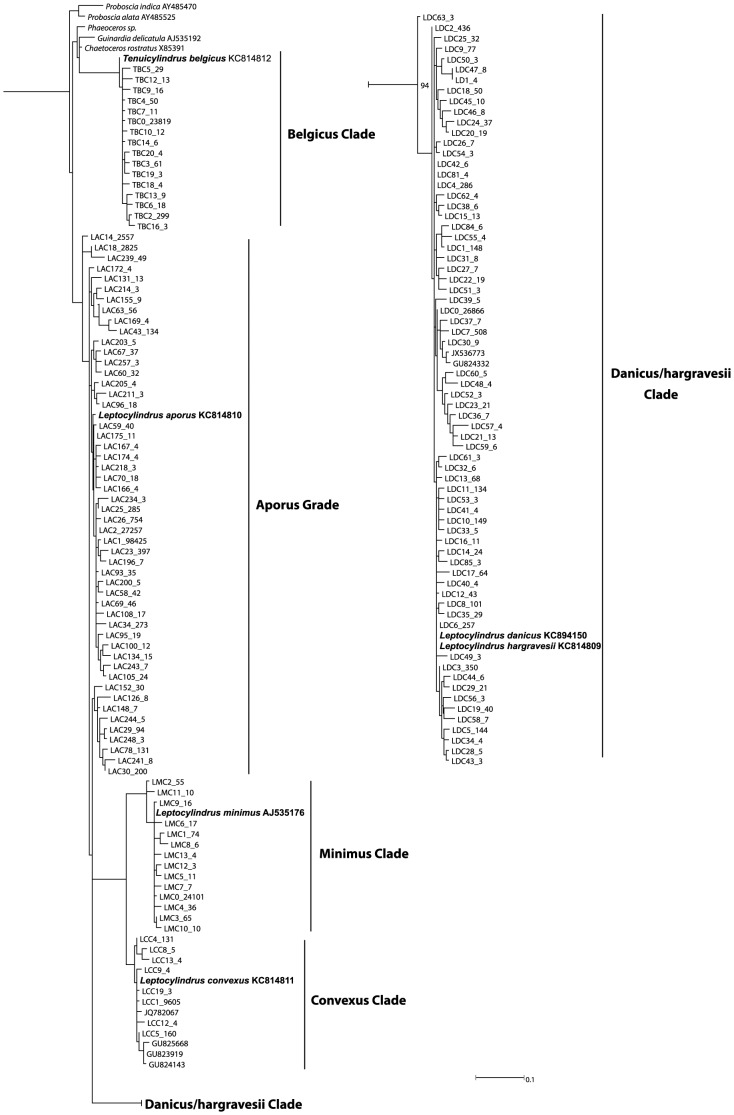
Figure 2. RAxML tree inferred from the alignment of 165 representative V9 sequences of leptocylindracean OTUs from the BioMarKs data, six leptocylindracean sequences from GenBank, and 96 reference sequences of bolidomonads, leptocylindraceans and other diatoms, utilizing the GTRGAMMA base substitution model and Hill Climbing algorithm.
Bolidomonas pacifica and B. mediterranea were designated as outgroups. All non-leptocylindracean sequences were pruned from the tree following tree construction (see Figure S2 for tree with outgroups included). Bootstrap values were inferred from 100 distinct alternative runs and values <50 are deleted. OTU labels follow same principle as in Figure 1.