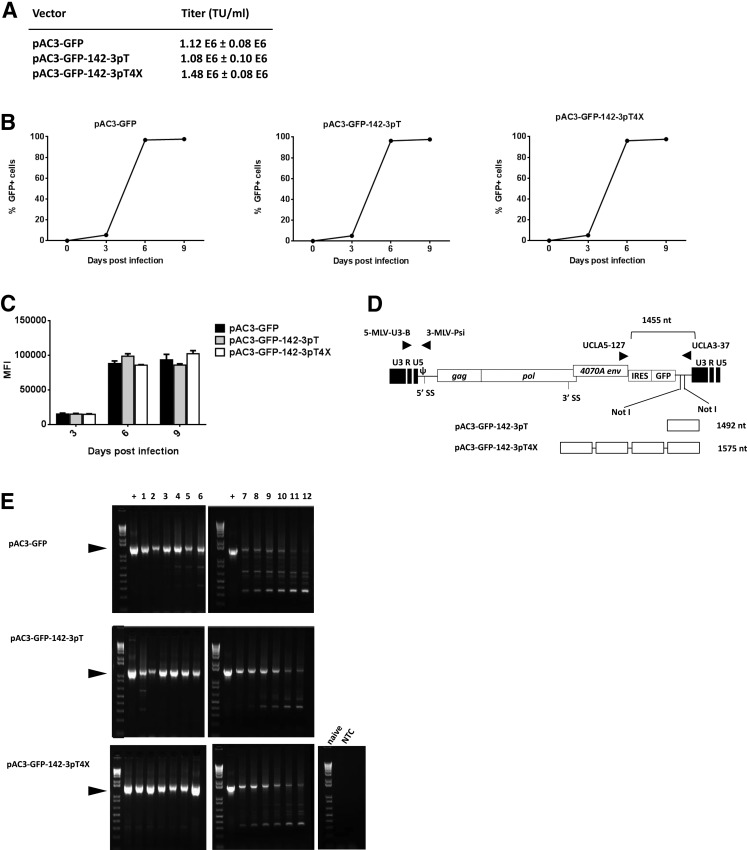
FIG. 2.
Replication kinetics and green fluorescent protein (GFP) expression levels of pAC3-GFP, pAC3-GFP-142-3pT, and pAC3-GFP-142-3pT4X vectors in U87-MG cells. (A) Viral titer of pAC3-GFP, pAC3-GFP-142-3pT, and pAC3-GFP-142-3pT4X vectors in PC-3 cells. (B) Replication kinetics of pAC3-GFP, pAC3-GFP-142-3pT, and pAC3-GFP-142-3pT4X vectors. U87-MG cells were infected with each vector at a multiplicity of infection (MOI) of 0.01 on day 0 and passaged on days 3, 6, and 9 postinfection. The percentage of GFP-positive cells was determined by flow cytometry with proper gating to exclude GFP-negative cells. The replication kinetics of each vector was obtained by plotting the percentage of GFP-positive cells versus time. (C) Comparison of MFI of GFP expression at the indicated time points postinfection. All experiments were performed in triplicate, and the data shown represent one of the three independent experiments (means+SD). (D) Schematic diagram of integrated proviral DNA and locations of the primer sets. 5-MLV-U3-B and 3-MLV-Psi primers and probe were used to determine the average copy number of vector per cell and viral titer by qPCR. UCLA5-127 and UCLA3-37 primers were used to assess the integrity of the IRES-GFP transgene of integrated proviral DNA spanning the IRES-GFP region by end-point PCR. (E) Stability of IRES-GFP transgene in pAC3-GFP, pAC3-GFP-142-3pT, and pAC3-GFP-142-3pT4X proviral DNA over multiple serial infections in U87-MG cells. DNA molecular marker (1 Kb Plus marker; Life Technologies) was included in the first lane of each gel. The numbers above each lane indicate the number of infection cycles for each vector. Arrowheads indicate size of the PCR product expected for the undeleted IRES-GFP region (1445 bp for pAC3-GFP, 1492 bp for pAC3-GFP-142-3pT, and 1575 bp for pAC3-GFP-142-3pT4X vector). NTC, no-template control; +, positive control using plasmid DNA corresponding to each vector as a template in PCR.