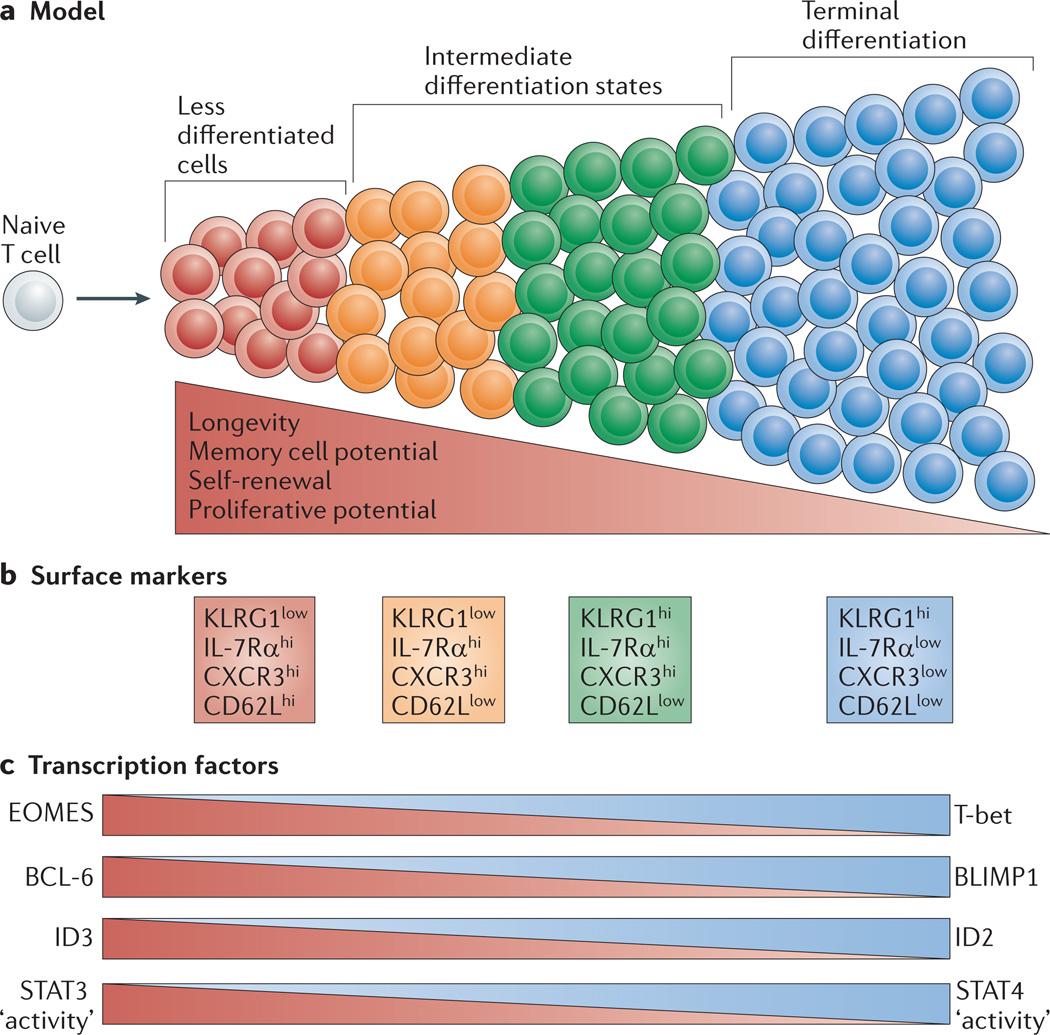
Figure 3. Graded activity of transcriptional programmes that control effector and memory T cell differentiation.
a | This model postulates that, in response to different levels of signal input, the differentiation of antigen-specific effector CD8+ T cells occurs along a continuum. Cells that have greater memory cell potential, longevity and proliferative potential are at one end of the spectrum, and terminally differentiated effector T cells are present at the other end of the spectrum. In between these two extreme end points are effector T cells that exist in intermediate differentiation states. b | The heterogeneous differentiation states of effector CD8+ T cells can be distinguished by the expression of several surface markers, such as killer cell lectin-like receptor G1 (KLRG1), IL-7 receptor subunit-α (IL-7Rα), CD27, CXC-chemokine receptor 3 (CXCR3) and CD62L. c | The transcriptional programmes that control terminal effector cell differentiation and memory cell potential seem to be based on the graded expression or activity of certain competing sets of transcription factors. As the relative expression or activity of T-bet, B lymphocyte-induced maturation protein 1 (BLIMP1), inhibitor of DNA binding 2 (ID2) and signal transducer and activator of transcription 4 (STAT4) increases and surpasses a given threshold during infection, effector CD8+ T cells acquire more terminally differentiated phenotypes that are associated with a reduction in proliferative capacity and longevity. These factors counter-regulate, and can also interact with, an opposing set of transcriptional regulators, including eomesodermin (EOMES), B cell lymphoma 6 (BCL-6), ID3 and STAT3, that prevent terminal differentiation of effector T cells and help to maintain memory cell properties. Note that to some extent this figure is a conceptual diagram, as the exact amounts of the different transcription factors or their activities have not been accurately measured in all cases.