Figure 4.
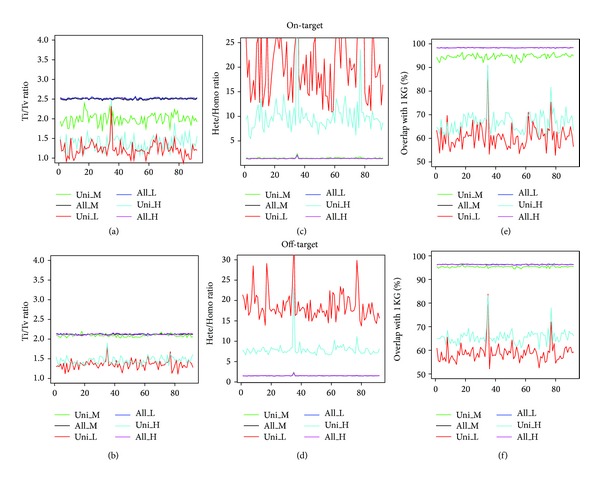
Comparison of the accuracy of SNPs detected by three strategies. The upper panel is plotted for SNPs across the on-target regions; the lower panel is for the off-target SNPs. From the left to right in each panel are the Ti/Tv ratio ((a) and (b)), Hete/Homo ratio ((c) and (d)), and overlapping rate with SNPs observed in the 1 KG project ((e) and (f)). Uni_M, Uni_L, and Uni_H represent the number of SNPs uniquely identified by the merged, low depth, and high depth strategies, respectively; All_M, All_L, and All_H denote the total number of SNPs identified by the merged, low depth, and high depth strategies, respectively. The x-axis represents each of 92 exome-seq samples.
