Abstract
Plastids utilize a complex gene expression machinery, which has coevolved with the underlying genome sequence. Relatively, little is known about the genome-wide evolution of transcript processing in algal plastids that have undergone complex endosymbiotic events. We present the first genome-wide study of transcript processing in a plastid acquired through serial endosymbiosis, in the fucoxanthin-containing dinoflagellate Karlodinium veneficum. The fucoxanthin dinoflagellate plastid has an extremely divergent genome and utilizes two unusual transcript processing pathways, 3′-poly(U) tail addition and sequence editing, which were acquired following the serial endosymbiosis event. We demonstrate that poly(U) addition and sequence editing are widespread features across the Karl. veneficum plastid transcriptome, whereas other dinoflagellate plastid lineages that have arisen through independent serial endosymbiosis events do not utilize either RNA processing pathway. These pathways constrain the effects of divergent sequence evolution in fucoxanthin plastids, for example by correcting mutations in the genomic sequence that would otherwise be deleterious, and are specifically associated with transcripts that encode functional plastid proteins over transcripts of recently generated pseudogenes. These pathways may have additionally facilitated divergent evolution within the Karl. veneficum plastid. Transcript editing, for example, has contributed to the evolution of a novel C-terminal sequence extension on the Karl. veneficum AtpA protein. We furthermore provide the first complete sequence of an episomal minicircle in a fucoxanthin dinoflagellate plastid, which contains the dnaK gene, and gives rise to polyuridylylated and edited transcripts. Our results indicate that RNA processing in fucoxanthin dinoflagellate plastids is evolutionarily dynamic, coevolving with the underlying genome sequence.
Keywords: dinoflagellate, chloroplast, RNA processing, editing, polyuridylylation, minicircle
Introduction
Plastid gene expression involves a complex set of transcriptional and posttranscriptional events. Some of the features of plastid gene expression, such as the use of a bacterial RNA polymerase and transcript cleavage, are likely to occur universally across the photosynthetic eukaryotes (Green 2011). Others, such as transcript splicing, sequence editing, and 3′-tail addition, appear to have evolved independently within individual plastid lineages (Asakura et al. 2008; Lange et al. 2009; Fujii and Small 2011), and this may be related to the evolution of the underlying genome sequence. For example, transcript editing in plant plastids, which is predominantly involved in cytosine deamination, is believed to have coevolved with an enrichment in the GC-content of the underlying genome sequence relative to the plastids of related green algae (Fujii and Small 2011).
Until recently, very little was known about the evolution of plastid transcript processing in lineages other than plants. Some of the most important emerging models for studying plastid gene expression in algae are dinoflagellates, and their closest relatives, such as the chromerid species Chromera velia and Vitrella brassicaformis (Howe et al. 2008; Janouskovec et al. 2013; Dorrell et al. 2014). Dinoflagellates are an evolutionarily diverse group of algae and nonphotosynthetic protists, and have important roles as free-living primary producers, and as symbionts of marine invertebrates such as coral (Howe et al. 2008). The ancestors of all extant dinoflagellates possessed a plastid of red algal origin, of the same endosymbiotic derivation as the plastids found in chromerids, which is retained in species that contain the pigment peridinin (Shalchian-Tabrizi et al. 2006; Janouskovec et al. 2010). The peridinin dinoflagellate plastid has an extremely reduced genome, containing fewer than 15 genes, many of which are highly divergent in sequence, and are located on small, plasmid-like elements termed “minicircles” (Zhang et al. 1999; Howe et al. 2008; Green 2011). Some dinoflagellates have replaced the peridinin-containing plastids with others of a different phylogenetic derivation, through serial endosymbiosis. For example, the fucoxanthin-containing dinoflagellates possess serially acquired plastids derived from haptophyte algae (Takishita et al. 1999; Gabrielsen et al. 2011; Dorrell and Howe 2012). A near-complete plastid genome sequence has been determined for the fucoxanthin dinoflagellate Karlodinium veneficum, which retains fewer genes than the plastids of free-living haptophytes, and is highly divergent in sequence (Gabrielsen et al. 2011; Espelund et al. 2012). Other serial endosymbiosis events have occurred in the dinoflagellate genus Lepidodinium, which possesses green algal plastids (Takishita et al. 2008; Matsumoto et al. 2011), and the “dinotom” algae, which possess plastids derived from diatoms (Takano et al. 2008; Imanian et al. 2010). Plastid genome sequences have been assembled for the dinotom species Kryptoperidinium foliaceum and Durinskia baltica, and these retain far more genes, and are less divergent in content than the Karl. veneficum plastid genome (Imanian et al. 2010; Gabrielsen et al. 2011).
In addition to possessing very unusual genomes, dinoflagellate plastids utilize a distinctive set of transcript processing pathways. Peridinin dinoflagellate plastid transcripts receive a 3′-terminal poly(U) tail, and this process, while also found in the plastids of chromerid algae, is absent from other plastid lineages, including those of haptophytes and diatoms (Wang and Morse 2006; Janouskovec et al. 2010; Dorrell and Howe 2012). In addition, plastid transcripts in some peridinin dinoflagellates undergo substitutional sequence editing, which can occur on up to one in ten residues in individual transcript sequences and has evolved independently from the much less extensive substitutional editing observed in land plant plastids (Zauner et al. 2004; Fujii and Small 2011; Dorrell and Howe 2012). Recently, we have shown that 3′-terminal poly(U) tail addition and sequence editing occur in plastids of the fucoxanthin dinoflagellate Karenia mikimotoi (Dorrell and Howe 2012). Editing has been demonstrated in Karl. veneficum, although poly(U) tails have not yet been reported (Jackson et al. 2013). As these pathways are associated with peridinin dinoflagellate plastids and are not found in free-living haptophytes, they are likely to be remnants of the ancestral peridinin-containing plastid symbiosis, applied to the fucoxanthin plastid following its uptake by the dinoflagellate host (Dorrell and Howe 2012). These very recently acquired transcript processing pathways in the highly divergent fucoxanthin dinoflagellate plastid provide a unique opportunity to explore the coevolution of plastid genes and gene expression pathways.
We have surveyed the distribution of poly(U) addition and editing sites across the entire published Karl. veneficum plastid genome (Gabrielsen et al. 2011; Espelund et al. 2012). Our study represents the first genome-wide investigation of RNA processing in a plastid acquired by serial endosymbiosis. We demonstrate that almost every gene in the Karl. veneficum plastid can give rise to polyuridylylated and edited transcripts, including genes that are not found in the plastid of peridinin dinoflagellates. We demonstrate that the serially acquired plastids in Lepidodinium and dinotoms do not utilize either of the RNA processing pathways. We have additionally identified unusual roles for poly(U) addition and editing in highly divergent regions of the Karl. veneficum plastid genome. Poly(U) addition may enable the differentiation of functional mRNAs from transcripts of pseudogenes that have arisen through recent genome rearrangements, and editing is associated with fast-evolving sequences and in-frame insertions that have arisen recently in fucoxanthin dinoflagellate plastids. In certain cases, these pathways may have indirectly contributed to the evolution of highly divergent sequences, such as a novel 3′-extension to the atpA coding sequence (CDS) that is generated through transcript editing. Most significantly, we present the first complete sequence of an episomal minicircle in a serially acquired dinoflagellate plastid, which has evolved convergently to the minicircles found in peridinin dinoflagellate plastids and gives rise to a polyuridylylated and edited dnaK transcript. Our data reveal extensive and complex coevolutionary trends between the plastid genome sequence and transcript processing machinery of fucoxanthin dinoflagellates.
Results
Presence of Poly(U) Tails on Karl. veneficum Plastid Transcripts
We investigated whether transcripts in the Karl. veneficum plastid receive 3′-poly(U) tails, as in the related fucoxanthin dinoflagellate Kare. mikimotoi (Dorrell and Howe 2012). As described previously, we performed reverse transcriptions of Karl. veneficum total cellular RNA using an oligo-d(A) primer. We then performed PCR using the same oligo-d(A) primer as the PCR reverse primer, and forward primers specific to a representative selection of genes across the Karl. veneficum plastid genome (supplementary table S1, Supplementary Material online) (Gabrielsen et al. 2011). These included five photosynthesis genes (psbA, psbC, psbD, psaA, and rbcL) previously shown to contain poly(U) sites in Kare. mikimotoi (fig. 1, lanes 1–5) (Dorrell and Howe 2012). We additionally tested two plastid housekeeping genes (rpl6 and rps5) that have not been investigated in Kare. mikimotoi and are not present in peridinin plastid genomes (fig. 1, lanes 6 and 7), and a 603-bp open reading frame (ORF) located in a 1,636-bp previously unannotated region between the Karl. veneficum chlI and psbL genes that shows no homology to any previously annotated nucleotide or protein sequence, which we henceforth term ORF1 (fig. 1, lane 8) (Gabrielsen et al. 2011). For each gene tested, we obtained products with the reverse transcriptase polymerase chain reaction (RT-PCR). These products were sequenced and found to correspond to transcripts containing poly(U) sequences located within the 3′-UTR of the gene concerned. These sequences did not correspond to poly(T) tracts in the Karl. veneficum plastid genome, and hence are posttranscriptional modifications to the transcript sequence. Our data thus suggest that a wide variety of transcripts in the Karl. veneficum plastid receive poly(U) tails, including transcripts of genes that are not found in the plastids of peridinin dinoflagellates.
Fig. 1.
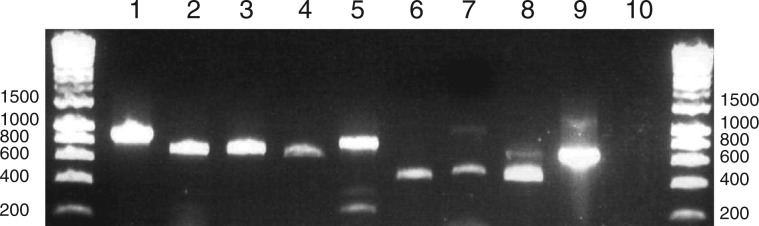
Presence of poly(U) tails in Karlodinium veneficum plastid transcripts. The gel photo shows the result of a series of representative oligo-d(A) RT-PCRs for specific transcripts from the Karl. veneficum plastid genome. Lanes 1–5: oligo-d(A) RT-PCRs of transcripts that have previously been shown to receive poly(U) tails in Karenia mikimotoi (psbA, psbC, psbD, psaA, rbcL). Lanes 6 and 7: oligo-d(A) RT-PCRs of representative housekeeping genes (rpl6, rps5). Lane 8: oligo-d(A) RT-PCR of the previously unannotated ORF1. Lane 9: RT-PCR of Karl. veneficum psbA using a cDNA template generated using an internal gene-specific cDNA synthesis and PCR reverse primer, and the same psbA forward primer used in lane 1. Lane 10: PCR using the same primers as lane 9, under template negative conditions. The faint secondary band at approximately 1,000 bp in lane 6 corresponds to a dicistronic polyuridylylated rpl6–rps5 transcript. The secondary bands visible in lanes 5, 8, and 9 were found to be PCR chimeras.
To confirm that the oligo-d(A) primed RT-PCR products correspond to 3′-terminal transcript poly(U) tails, as opposed to internal sequence insertions, or to artifacts generated by mispriming of the oligo-d(A) primer, we performed RT-PCRs using circular RNA and cDNA and PCR synthesis primers specific to the Karl. veneficum psbA and psbC genes (supplementary table S1, Supplementary Material online). We have previously employed this technique successfully to confirm the presence of polyuridylylated psbA and psbC transcripts in Kare. mikimotoi (supplementary fig. S1, Supplementary Material online) (Dorrell and Howe 2012). We identified 3′-terminal poly(U) tails on the ends of Karl. veneficum psbA and psbC transcripts using this approach (supplementary fig. S1, Supplementary Material online). Although we additionally identified nonpolyuridylylated psbA transcripts, all of these transcripts terminated at the 3′-end within the CDS and are therefore likely to represent transcript degradation products as opposed to mature transcripts generated by a poly(U)-independent processing pathway (supplementary fig. S1, Supplementary Material online). Our data confirm that poly(U) tails are added to a wide variety of plastid transcripts in Karl. veneficum, as with Kare. mikimotoi, and suggest that the poly(U) addition pathway was acquired by a common ancestor of extant fucoxanthin dinoflagellates.
Extent of Poly(U) Addition within the Karl. veneficum Plastid
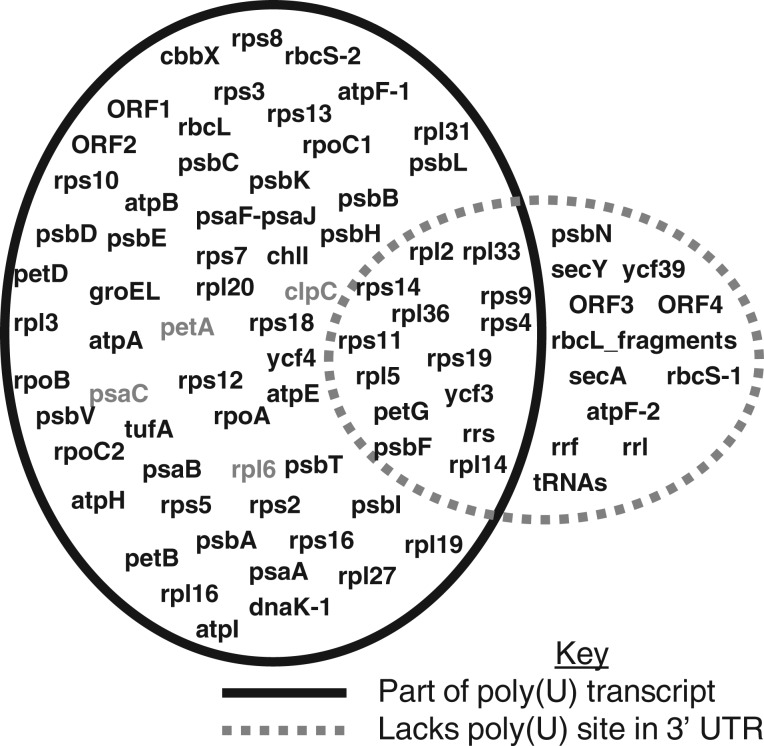
We extended the initial analysis to determine the total extent of transcript polyuridylylation in the Karl. veneficum plastid. We performed oligo-d(A) primed RT-PCRs using PCR forward primers for every annotated protein-coding and ribosomal RNA gene within the plastid genome, including previously unannotated atpE, petG, and rps10 genes (supplementary tables S1 and S2, Supplementary Material online). We also tested for the presence of poly(U) tails for 15 predicted tRNA genes in the Karl. veneficum plastid genome, and three further predicted ORFs of more than 300 bp length that bear no sequence homology to any previously identified plastid gene (supplementary table S1, Supplementary Material online) (Gabrielsen et al. 2011). We found evidence for widespread polyuridylylation of the Karl. veneficum plastid transcriptome, with 54 of the 75 protein-coding genes, and two of the four novel ORFs surveyed possessing poly(U) sites in the associated 3′-UTR (fig. 2 and supplementary table S2, Supplementary Material online). Four of the 56 poly(U) sites observed were positioned within genomic poly(T) tracts (supplementary table S2, Supplementary Material online), so it is possible that they have arisen through primer misannealing. However, the remaining 52 were not and are likely to correspond to posttranscriptional modifications.
Fig. 2.
Extent of transcript polyuridylylation across the Karlodinium veneficum plastid. The Venn diagram shows the transcript polyuridylylation state of every gene within the Karl. veneficum plastid genome. Genes in the overlap sector between the two circles lack directly associated poly(U) sites in their respective 3′-UTR sequences, but can be retrieved as part of polyuridylylated polycistronic transcripts, with the poly(U) site positioned in the 3′-UTR of a downstream gene. The poly(U) tails of genes shaded in gray may be generated by the transcription of genomic poly(T) tracts.
For some of the oligo-d(A) RT-PCRs we identified multiple products, consistent with the presence of different polyuridylylated transcripts from a single gene. For example, in the case of rpl6, in addition to obtaining PCR products of a size consistent with a monocistronic, polyuridylylated transcript, we observed a secondary higher molecular weight product that was found to correspond to a polyuridylylated dicistronic rpl6–rps5 transcript (fig. 1, lane 7). We additionally obtained polyuridylylated dicistronic transcripts for 13 of the 21 protein-coding genes that lacked poly(U) sites immediately downstream but were positioned directly upstream of genes that possessed poly(U) sites (supplementary table S2, Supplementary Material online, and fig. 2). This indicates that even genes that do not possess directly adjacent poly(U) sites may give rise to polyuridylylated transcripts.
A small number of the protein-coding genes and unannotated ORFs in the Karl. veneficum plastid failed to yield significant products in any oligo-d(A) RT-PCR attempted (supplementary table S2, Supplementary Material online, and fig. 2). In each case, we failed to detect products for each gene even following a nested reamplification of the primary PCR product, with the same oligo-d(A) primer and a second gene-specific primer positioned downstream of the first (supplementary table S1, Supplementary Material online). None of these genes was positioned directly upstream of a gene in the same transcriptional orientation that possessed a poly(U) site, suggesting that they are unlikely to give rise to polycistronic polyuridylylated transcripts (supplementary fig. S2 and table S2, Supplementary Material online). We amplified transcript sequences for these genes, using cDNA synthesis primers internal to the CDS, and these were not completely identical to the underlying genomic sequence, consistent with transcript editing (supplementary tables S1 and S2, Supplementary Material online). We similarly could not identify products in oligo-d(A) RT-PCRs using primers specific to any of the ribosomal RNA subunits or tRNA genes although we identified a tricistronic polyuridylylated rrs-petG-atpF-1 transcript (fig.2 and supplementary table S1, Supplementary Material online). We generated transcript sequences for all three ribosomal subunits (5.8S, 16S, and 23S rRNA), and could detect low levels of editing in each case (supplementary tables S1 and S2, Supplementary Material online). Our data indicate that the poly(U) addition and editing machinery have been co-opted to recognize almost every gene in the Karl. veneficum plastid.
Location of Poly(U) Sites
We wished to determine what sequence features were associated with the presence of poly(U) sites in the Karl. veneficum plastid genome. In chromerid algae, poly(U) addition is biased toward genes encoding proteins that function in photosynthesis (Dorrell et al. 2014). Although photosynthesis genes in the Karl. veneficum plastid are more likely to possess an associated poly(U) site than housekeeping genes, the association is not statistically significant (chi-squared, P = 0.07) (fig. 2 and supplementary table S2, Supplementary Material online). We additionally compared the gene order of the Karl. veneficum genome with those of free-living haptophyte species, and could not identify a consistent relationship between the absence of a poly(U) site, and inferred recombination events (supplementary table S3, Supplementary Material online). Our data therefore indicate that gene function and genome rearrangements are unlikely to be the only factors that determine the distribution of poly(U) sites across the Karl. veneficum plastid.
The poly(U) sites within the Karl. veneficum plastid are typically positioned close to the 3′-end of the CDS, with an average 3′-UTR length of only 30 bp (supplementary table S2, Supplementary Material online). We looked for conserved primary sequence motifs, changes in GC and purine/pyrimidine content, and predicted RNA secondary structures in the 3′-UTR sequences of each gene, extending 100 bp downstream of each poly(U) site. We could not identify any sequence features that were significantly associated with the presence of a poly(U) site. Several of the poly(U) sites, however, were located within the CDS of the downstream gene (supplementary table S2, Supplementary Material online). Most dramatically, within the ten-gene ribosomal protein operon extending from rpl3 through to rps5, we identified four genes (rpl3, rpl16, rps8, and rpl6) where the poly(U) site overlaps with the downstream CDS, whereas we only found one poly(U) site within a 3′-UTR sequence, associated with rps5 (supplementary fig. S3, Supplementary Material online) (Gabrielsen et al. 2011). Using a forward primer specific to rpl2, we additionally detected a poly(U) site located 296 bp within the rpl2 CDS although we could not identify this site using a forward primer specific to the upstream rpl3 gene (supplementary fig. S3, Supplementary Material online). The poly(U) sites located internal to gene sequences might be associated with alternative end processing events as their formation would prevent transcripts of specific genes being produced from polycistronic precursors. Overall, our data suggest that instead of poly(U) addition being associated with common sequence motifs or specific genes, poly(U) sites are highly sequence-specific. The formation of specific poly(U) sites might influence other events in plastid transcript processing.
Differential Recognition of Pseudogenes by the Karl. veneficum Plastid Transcript Processing Machinery
It has been demonstrated that poly(U) addition discriminates between paralogous copies of genes in the C. velia plastid (Dorrell et al. 2014). Transcripts of the C. velia atpH-1 gene, which are abundant, receive a poly(U) tail, whereas transcripts of the atpH-2 gene, which appears to be a pseudogene, do not (Janouskovec et al. 2013; Dorrell et al. 2014). Several of the genes in the Karl. veneficum plastid are present in multiple copies, some of which appear to be functional, whereas others are likely to be pseudogenes (Gabrielsen et al. 2011). For example, two copies of the rbcS gene are present: rbcS-2, which is likely to encode a functional protein, and rbcS-1, which contains an in-frame insertion within the region encoding the βC-βD loop domain of the rubisco small subunit, that if expressed would be likely to interfere with its function (supplementary fig. S4A, Supplementary Material online) (Larson et al. 1997; Li et al. 2005). Similarly, we identified two copies of the atpF gene: a previously annotated gene (atpF-1) and a previously unannotated pseudogene (atpF-2), positioned downstream of and in reverse orientation to psbB, which contains an internal frame-shift sequence deletion that would prevent the translation of the complete protein sequence (supplementary fig. S4B and table S2, Supplementary Material online).
We wished to determine whether transcripts of the rbcS-1 and atpF-2 pseudogenes receive poly(U) tails and are edited. We could detect polyuridylylated rbcS-2 and atpF-1 transcripts by oligo-d(A) primed RT-PCR, using PCR forward primers specific to each sequence (fig. 3, lanes 2 and 5), but could not detect polyuridylylated rbcS-1 and atpF-2 transcripts through the same approach (fig. 3, lanes 1 and 6). We could amplify nonpolyuridylylated rbcS-1 and atpF-2 transcript sequences by performing RT-PCRs against cDNA synthesis primers specific to each gene (fig. 3, lanes 3–4, 7–8). We sequenced the products of these RT-PCRs, and confirmed the presence of the in-frame insertion in rbcS-1 and the frame-shift deletion in atpF-2 (supplementary fig. S4, Supplementary Material online). We could not identify any editing within the atpF-2 transcript, and detected only one editing event on the rbcS-1 transcript, which is significantly fewer than the 15 editing events observed over the same region of the rbcS-2 transcript sequence (supplementary table S2, Supplementary Material online; binomial test, P< E-05). Our data indicate that poly(U) addition and editing are preferentially associated with functional genes in the Karl. veneficum plastid.
Fig. 3.
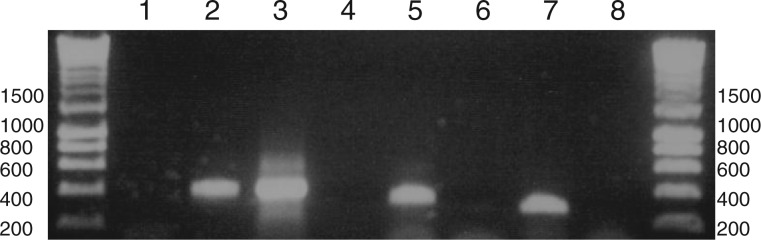
Specific addition of poly(U) tails to transcripts of functional gene paralogs in the Karlodinium veneficum plastid. This gel photo shows the result of a series of RT-PCRs to identify whether transcripts of functional and pseudogenic copies of the rbcS and atpF genes in the Karl. veneficum plastid receive poly(U) tails. Lanes 1 and 2: oligo-d(A) RT-PCR of rbcS-1 (pseudogene) and rbcS-2 (functional). Lanes 3 and 4: RT-PCR of rbcS-1 with a gene-specific internal cDNA synthesis primer under template positive (lane 3) and negative (lane 4) conditions. Lanes 5 and 6: oligo-d(A) RT-PCR of atpF-1 (highly divergent gene) and the atpF-2 nonfunctional sequence between rps16 and psbB. Lanes 7 and 8: RT-PCR of the atpF-2 region with a gene-specific cDNA synthesis primer under template positive (lane 7) and negative (lane 8) conditions.
Global Trends in Editing across the Karl. veneficum Plastid Transcriptome
Recently, Jackson et al. (2013) have profiled editing events in the Karl. veneficum plastid by comparing transcript and genomic sequences for regions of 14 different genes. Four different forms of editing were observed, all of which were transitions, consisting predominantly of A to G and U to C editing events, as well as small numbers of G to A and C to U conversions (Jackson et al. 2013). Across our entire data set, we found evidence for extensive sequence editing (table 1 and supplementary table S2, Supplementary Material online). Approximately 4.3% of sites in our transcript sequences were edited, slightly higher than previous estimates (Jackson et al. 2013). For some genes, we detected higher frequencies of editing, extending to 14% of positions for the Karl. veneficum psbD gene, and 24% of residues in the highly divergent petG sequence (supplementary table S2, Supplementary Material online). Editing sites were situated predominantly within gene sequences although we detected a low level of editing (1.6%) in polyuridylylated transcript 3′-UTR sequences (supplementary table S2, Supplementary Material online), as previously seen in Kare. mikimotoi (Dorrell and Howe 2012). Many (88%) of the editing events lead to an increase in transcript GC-content, consistent with previous studies (Dorrell and Howe 2012; Jackson et al. 2013) (table 1). Although the majority (96%) of editing events observed were transition events, we detected seven different transversion events at low levels in the Karl. veneficum transcriptome, similar to our previous observations in Kare. mikimotoi (table 1) (Dorrell and Howe 2012).
Table 1.
Total Editing Events from the Characterized Plastid Transcriptomes of Karenia mikimotoi and Karlodinium veneficum.
| Overview | Karenia | Karlodinium—Jackson | Karlodinium—Extended |
|---|---|---|---|
| Sequence length | 5,473 | 7,373 | 36,084 |
| A-C | 26 | 0 | 15 |
| A-G | 59 | 131 | 789 |
| A-U | 0 | 0 | 16 |
| C-A | 1 | 0 | 8 |
| C-G | 0 | 0 | 4 |
| C-U | 17 | 8 | 49 |
| G-A | 15 | 15 | 99 |
| G-C | 24 | 0 | 8 |
| G-U | 0 | 0 | 1 |
| U-A | 0 | 0 | 11 |
| U-C | 116 | 67 | 540 |
| U-G | 2 | 0 | 11 |
| Total | 260 | 221 | 1,539 |
| % Bases edited | 4.75 | 2.86 | 4.27 |
| % Transitions | 79.6 | 100 | 96 |
| % Transversions | 20.4 | 0 | 4 |
| % GC-enrich | 78.1 | 89.6 | 88 |
| % GC-deplete | 12.7 | 10.4 | 10 |
| % GC-neutral | 9.2 | 0 | 1.9 |
| % Nonsynonymous | 58.5 | 95 | 87.1 |
| % Synonymous | 41.5 | 5 | 12.9 |
Note.—The total editing events observed across 36,084-bp Karlodinium veneficum plastid transcript sequence in this study are profiled, alongside previous surveys of Karl. veneficum (Jackson et al. 2013), and of the related fucoxanthin dinoflagellate Kare. mikimotoi (Dorrell and Howe 2012).
Most (87%) of the editing events in the Karl. veneficum plastid are predicted to have nonsynonymous effects on the corresponding protein sequence (table 1). Some of these editing events may be required for the correct function of the encoded protein. For example, 11 of the genes in the Karl. veneficum plastid contain premature in-frame termination codons, which would prevent the translation of the complete protein sequence. Correction of premature termination codons through editing has previously been reported for Karl. veneficum rpoB, rps13, psaA and secY transcripts, and psaA in Kare. mikimotoi (Dorrell and Howe 2012; Jackson et al. 2013). We confirmed that all of the premature termination codons in the Karl. veneficum genome are removed from the corresponding polyuridylylated transcript sequences by editing (supplementary table S4, Supplementary Material online). Consistent with previous reports, we also found that edited Karl. veneficum transcripts show an increase in sequence similarity, relative to the genomic sequence, to the corresponding sequences from the haptophytes Emiliania huxleyi and Phaeocystis globosa (supplementary table S4, Supplementary Material online) (Jackson et al. 2013). Editing in the Karl. veneficum plastid therefore appears to reduce the effects of divergent mutations on plastid protein sequence.
Editing of Fast-Evolving Sequences in the Karl. veneficum Plastid
Not all of the nonsynonymous editing events observed within the Karl. veneficum plastid have readily inferred effects on plastid protein function. Across our entire data set, we found that although more than one in ten codons undergo a nonsynonymous change due to editing, this only leads to a net increase of 1.6% in sequence conservation between the Karl. veneficum and haptophyte protein sequences (supplementary table S4, Supplementary Material online). The other editing events may have selectively neutral or disadvantageous effects, or affect sequences that are not found in free-living haptophytes. Notably, many of the genes in the Karl. veneficum plastid genome contain novel sequence insertions, or fast-diverging regions that bear no homology to haptophyte sequences (Gabrielsen et al. 2011). We hypothesized that editing events that do not increase sequence conservation with haptophyte orthologs might instead affect sequences unique to the Karl. veneficum plastid.
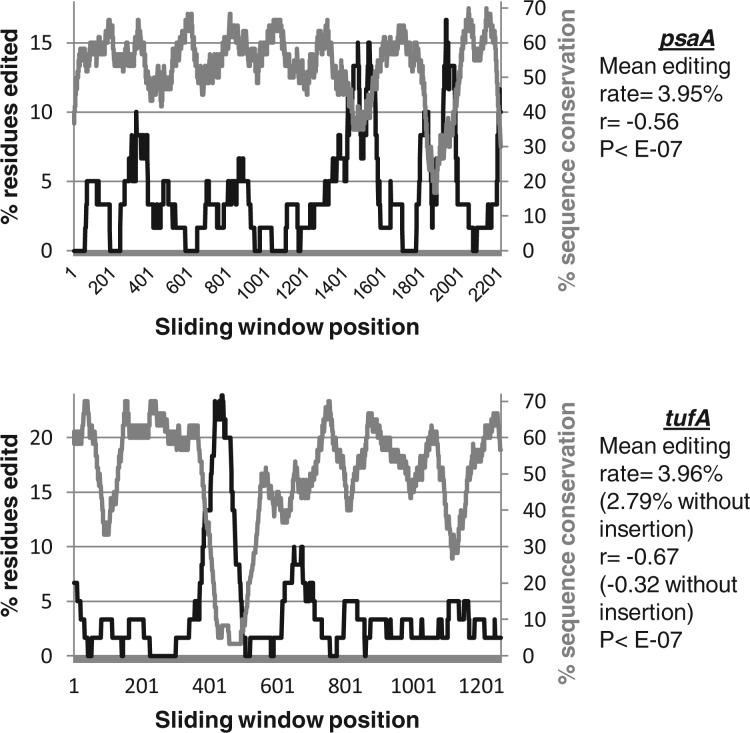
Certain transcripts within our data set contain highly edited regions. For example, the psaA and tufA genes both contain small regions where >15% of residues are edited, compared with an average editing rate across each gene of approximately 4% (fig. 4). To test whether these highly edited sites correspond to particularly divergent sequences, we calculated editing frequencies using a sliding 60-bp window, in polyuridylylated transcripts covering the entire psaA and tufA gene sequences. We additionally calculated the predicted sequence conservation, over the same sliding window, between the predicted Karl. veneficum psaA and tufA transcript translation products, and the corresponding E. huxleyi protein sequences (fig. 4). In both genes, editing was specifically correlated with low sequence conservation with E. huxleyi (Pearson correlation = −0.56 for psaA, −0.67 for tufA; P < E-07 for both genes). Notably, over a third of the editing events within tufA occur within an 84-bp region, which forms less than one-twelfth of the entire gene and is significantly more highly edited than the rest of the sequence (chi-squared: P < 0.05). This region corresponds to an in-frame insertion unique to Karl. veneficum (supplementary fig. S5, Supplementary Material online). Overall, our data indicate that editing events are associated with regions of sequence that are recently acquired or are highly divergent. Editing might reduce the effects of these divergent sequences on protein function.
Fig. 4.
Editing is preferentially associated with highly divergent regions of Karlodinium veneficum plastid genes. These graphs compare the frequency of editing with sequence conservation in a 60-bp sliding window over the entire lengths of the Karl. veneficum psaA and tufA genes. The horizontal axis shows the starting position of each window within each gene sequence. The left hand vertical axis of each graph (black line) depicts the total percentage of nucleotide positions within each window that are edited within the transcript sequence. The right hand vertical axis (gray line) shows the proportion of amino acid positions within the predicted translation product of the transcript sequence of this window that are conserved with the predicted translation of the orthologous gene in the Emiliania huxleyi plastid. A table to the right-hand side of each graph shows the total proportion of editing sites over the entire gene, and the Pearson coefficient, and associated significance value of the correlation between sequence conservation and editing frequency. For the tufA gene, correlation coefficients are given both for the complete gene sequence (open figures) and for the gene sequence excluding the highly edited 84-bp insertion region specific to Karl. veneficum (bracketed figures). In all cases, a significant negative correlation is observed.
Editing-Facilitated Divergent C-Terminal Evolution of Karl. veneficum AtpA
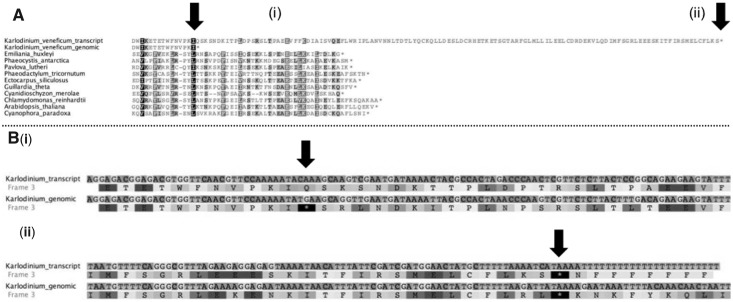
For the Karl. veneficum atpA gene, editing appears to be involved in the generation of a novel 3′-extension on the conventional CDS (fig. 5). The Karl. veneficum atpA gene contains a premature in-frame TGA codon, which is edited to form a CAA-glutamine codon in the mature transcript sequence. However, the Karl. veneficum atpA gene does not contain the consensus 3′-end found in other plastid sequences. The translation product of the Karl. veneficum atpA transcript is similar in sequence up to the final six amino acids in the E. huxleyi plastid AtpA protein, where it diverges to contain a 95-aa C-terminal extension that bears no homology to any other known sequence (fig. 5). The expression of this extension would be possible only from edited transcript sequence, and therefore transcript editing may have enabled divergent evolution of the ATP synthase complex in the Karl. veneficum plastid.
Fig. 5.
Generation of a novel C-terminal sequence extension by editing of Karlodinium veneficum atpA transcripts. (A) An alignment of the predicted translation products of the genomic and transcript sequences of Karl. veneficum atpA with protein sequences from other plastid lineages. (B) A nucleotide sequence alignment, and predicted translation products of two regions of the Karl. veneficum genomic and transcript sequence in detail. Residues important for defining the size of the predicted translation product of each Karl. veneficum sequence are labeled with vertical arrows. The Karl. veneficum genomic translation product terminates approximately 33-aa upstream of the consensus AtpA C-terminus, due to the presence of an in-frame TGA STOP codon within the atpA gene sequence. This is altered by editing to a CAA-Gln codon (B[i]) in the transcript sequence, enabling the translation of the complete AtpA C-terminus. However, the atpA transcript sequence is highly divergent at the 3′-end, and does not possess a termination codon at the consensus position relative to orthologous AtpA sequences. Instead, it encodes an 85-aa extension sequence that is not conserved with other AtpA sequences, which terminates in an unedited TAA STOP codon (B[ii]).
Expression and Transcript Processing of Minicircles Located in the Karl. veneficum Plastid
Certain genes within the Karl. veneficum plastid genome, such as rbcL and dnaK, are enriched in sequencing libraries relative to others (Espelund et al. 2012). These genes have been shown not only to be encoded on the chromosomal Karl. veneficum plastid genome sequence but also on multiple small elements, containing fragments of individual genes, that do not assemble onto the plastid genome (Espelund et al. 2012).The episomal elements have been suggested to correspond to a population of plastid-located minicircles, which have arisen independently of those found in peridinin dinoflagellates (Zhang et al. 1999; Howe et al. 2008; Espelund et al. 2012). However, it is not known whether these episomal elements are located in the Karl. veneficum plastid, nor has a complete episomal element yet been sequenced and confirmed to form a minicircle.
We investigated whether episomal fragments in Karl. veneficum may give rise to polyuridylylated transcripts. Polyuridylylation is not found in dinoflagellate nuclei or mitochondria and would accordingly confirm localization of the elements to the Karl. veneficum plastid (Dorrell and Howe 2012). We initially designed primers specific to the chromosomal and episomal copies of rbcL and tested for the presence of polyuridylylated transcripts by oligo-d(A) primed RT-PCR, as before, but could not identify any evidence for poly(U) addition or editing on transcripts of the episomal rbcL elements, in contrast to transcripts of the chromosomal rbcL gene (supplementary fig. S6, Supplementary Material online).
We additionally investigated the transcription of episomal dnaK genes. Although there is a complete copy of the rbcL gene within the Karl. veneficum plastid genome, the chromosomal dnaK genes lack consensus terminal regions and contain frame-shift mutations, suggesting that they do not give rise to translationally functional dnaK transcripts (supplementary fig. S7A, Supplementary Material online) (Gabrielsen et al. 2011; Espelund et al. 2012). We could not identify polyuridylylated transcripts from either chromosomal dnaK gene. Instead, using PCR primers designed against different regions of dnaK sequence, we identified a single polyuridylylated transcript, which we term dnaK-1 (supplementary fig. 7A and supplementary table S1, Supplementary Material online). The dnaK-1 transcript encodes a complete plastid Hsp70 and does not contain any frame-shifts or align with either chromosomal dnaK gene, suggesting that it is expressed from an episomal element.
To identify what genetic elements might give rise to the dnaK-1 transcript, we performed thermal asymmetric interlaced PCR (Liu et al. 1995), using combinations of primers derived from the dnaK-1 transcript sequence. We identified a single gene that covered the entire dnaK-1 CDS and 3′-UTR past the poly(U) site. The dnaK-1 poly(U) site coincides with a genomic T12 motif; however, we identified dnaK-1 transcripts through circular RT-PCR with poly(U) tails of up to 19 nt length, implying that they are generated through posttranscriptional sequence modification (supplementary fig. S7B, Supplementary Material online). In addition, we found evidence of extensive editing in the dnaK-1 transcript sequence (supplementary table S2, Supplementary Material online). Overall, our data imply that dnaK-1 is transcribed from a single contiguous genetic element, located within the Karl. veneficum plastid, but separate from the chromosomal genome sequence.
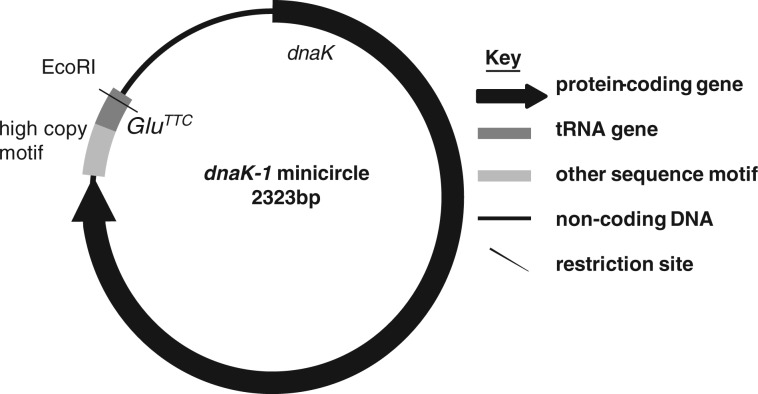
Surprisingly, the dnaK-1 3′-UTR obtained was found to extend into a region of sequence identical to the 5′-end of the dnaK-1 gene, consistent with the dnaK-1 gene being located on a plastid minicircle (fig. 6). The dnaK-1 minicircle is 2,323 bp long and contains a single EcoRI restriction site, which is consistent with a 2.3-kbp band containing the dnaK gene identified through Southern blotting of EcoRI-digested Karl. veneficum gDNA (fig. 6) (Espelund et al. 2012). In addition to a complete dnaK gene, this minicircle contains a GluTTC tRNA gene, and a single “high copy” region that is conserved with other episomal sequences previously identified from Karl. veneficum (fig. 6) (Espelund et al. 2012). This is the first complete plastid minicircle identified in a fucoxanthin dinoflagellate, confirming that the fucoxanthin plastid genome has undergone a similar fragmentation to that observed in peridinin dinoflagellate plastid genomes. Our data furthermore show that the poly(U) and editing machinery of fucoxanthin dinoflagellates may recognize transcripts of genes encoded on minicircles over genes located on the chromosomal plastid genome.
Fig. 6.
Schematic diagram of the Karlodinium veneficum dnaK-1 minicircle. The 2,323-bp dnaK-1 minicircle contains a complete dnaK-1 positioned directly upstream of the predicted “high copy element,” and a GluTCC tRNA gene in the same transcriptional orientation. A single EcoRI restriction site is present on the reverse strand of the tRNA gene.
Absence of Poly(U) Addition and Editing from Diatom and Green Algal-Derived Serially Acquired Dinoflagellate Plastids
We wished to determine whether poly(U) addition and transcript editing are found in either dinotom or green dinoflagellate plastids, as in the fucoxanthin and peridinin-containing lineages. We performed oligo-d(A) primed RT-PCRs on five genes (psbA, psbC, psbD, psaA, and rbcL) using total cellular RNA, and PCR primers specific to the dinotom Kryptoperidinium foliaceum and green dinoflagellate Lepidodinium chlorophorum (supplementary fig. S8A, Supplementary Material online). We could not detect polyuridylylated transcripts for any of the genes tested (supplementary fig. S8A, lanes 1–5, 7–11, Supplementary Material online). We detected nonpolyuridylylated psbA transcripts in both species using gene-specific cDNA synthesis primers (supplementary fig. S8A, lanes 6 and 12, Supplementary Material online) and by circular RT-PCR (supplementary fig. S8B, Supplementary Material online). We could not find any evidence of editing on these transcript sequences. We conclude that poly(U) addition and editing are found only in dinoflagellates that possess the ancestral peridinin plastid or the fucoxanthin replacement lineage.
Discussion
We have characterized the distribution and function of transcript editing and poly(U) tail addition across the entire plastid genome of the fucoxanthin dinoflagellate Karl. veneficum. This represents the first genome-wide study of transcript processing in a plastid acquired through serial endosymbiosis. The demonstration of poly(U) addition in Karlodinium, as in Kare. mikimotoi, indicates that it was acquired by a common ancestor of all studied fucoxanthin dinoflagellates (Bergholtz et al. 2006; Gabrielsen et al. 2011). We also found extensive sequence editing events, consistent with previous studies that identified them in both fucoxanthin dinoflagellate species (Dorrell and Howe 2012; Jackson et al. 2013). These editing events include transversion substitutions that have not previously been detected in Karl. veneficum but do occur in Kare. mikimotoi, suggesting that these are conserved across extant fucoxanthin dinoflagellates (Dorrell and Howe 2012; Jackson et al. 2013).
Many of the features associated with poly(U) addition and editing in Karl. veneficum have previously been documented in peridinin dinoflagellates. Multiple types of editing events have already been observed in peridinin dinoflagellates, and all species studied have had overall rates of editing of under 5%. In all species, A-G and U-C editing have been the two most abundant types of editing event (Zauner et al. 2004; Wang and Morse 2006; Dang and Green 2009; Iida et al. 2009; Mungpakdee et al. 2014). As in peridinin dinoflagellates, almost every protein-coding gene within the Karl. veneficum plastid can give rise to polyuridylylated transcripts, whereas tRNA genes do not possess poly(U) sites (Wang and Morse 2006; Nelson et al. 2007; Barbrook et al. 2012). Similarly, polyuridylylated polycistronic transcripts and poly(U) sites that overlap with adjacent gene sequences have previously been identified in peridinin dinoflagellates and in chromerids (Barbrook et al. 2012; Dorrell et al. 2014). This suggests that poly(U) addition has a similar functional role in transcript processing in both peridinin and fucoxanthin dinoflagellate plastids.
We also identified properties of poly(U) addition and editing that are specific to fucoxanthin dinoflagellate plastids. The editing events in Karl. veneficum include C-A, C-G, and U-A editing events that have not previously been detected in any peridinin dinoflagellate species although C-A editing has also been detected in Kare. mikimotoi (Dorrell and Howe 2012). Many of the poly(U) sites within the Karl. veneficum plastid are associated with housekeeping genes, which are not retained in the plastid genomes of peridinin dinoflagellates (Bachvaroff et al. 2004; Howe et al. 2008), and are plastid-located but typically do not possess poly(U) sites in chromerids (Dorrell et al. 2014).
Other unusual transcript processing features are associated with particularly divergent sequences in the Karl. veneficum plastid genome. The absence of poly(U) sites associated with pseudogenes has been described in chromerids (Janouskovec et al. 2013; Dorrell et al. 2014), but neither this nor a difference in the frequency of editing events on functional versus pseudogene transcripts has previously been reported in peridinin dinoflagellates. In contrast, at least some pseudogene transcripts in peridinin dinoflagellates are known to be extensively edited (Iida et al. 2009). Poly(U) addition and editing might therefore have a role in discriminating functional genes from nonfunctional gene fragments generated by recent rearrangements in fucoxanthin dinoflagellate plastid genomes. Similarly, the association of editing sites with fast-evolving sequences, such as the in-frame insertion in tufA, has not been described in other dinoflagellates and contrasts with plastid editing in plants, which is predominantly associated with slowly evolving sites within the genome sequence (Fujii and Small 2011; Hayes et al. 2012). These editing events might help neutralize the effects of fast-diverging sequences and recently acquired insertions on protein function.
In other cases, our data indicate that editing and poly(U) addition may have indirectly facilitated divergent sequence evolution in fucoxanthin dinoflagellate plastids. Sequence editing may have permitted the establishment of a novel 3′-sequence extension on transcripts of the Karl. veneficum atpA gene. To our knowledge, the edited extension of a plastid transcript into nonconserved sequence has never previously been reported. Most significantly, we have identified one plastid gene—dnaK—for which polyuridylylated and edited transcripts are derived from an episomal minicircle. This represents the first complete plastid minicircle sequence from a fucoxanthin dinoflagellate and suggests that the plastid genomes of fucoxanthin and peridinin dinoflagellates are undergoing convergent evolution events (Zhang et al. 1999; Espelund et al. 2012). The preferential targeting of the poly(U) and editing to dnaK gene copies located on minicircles may have led to their fixation over copies located on the chromosomal plastid genome, which appear to have been reduced to pseudogenes (Gabrielsen et al. 2011; Espelund et al. 2012).
Overall, our data indicate that poly(U) addition and editing in Karl. veneficum have evolved dynamically alongside the underlying genome, reducing the effects of mutations on plastid function, and potentially enabling the evolutionary fixation of divergent sequences. It remains to be seen whether poly(U) addition and editing were acquired after the extremely fast sequence evolution observed in the Karl. veneficum plastid commenced, or whether fucoxanthin plastid genomes and transcript processing have a more tightly interconnected evolutionary history. This might be resolved by investigating genome and transcriptome evolution in Kare. mikimotoi, or other less well-characterized fucoxanthin dinoflagellate plastids (Takishita et al. 1999; Bergholtz et al. 2006). Notably, the serially acquired plastids of dinotoms, which have less divergent genome sequences than fucoxanthin dinoflagellates, and of Lepidodinium, do not apply poly(U) tails or edit plastid transcripts. It will be worth determining whether the dinotoms, or Lepidodinium, have retained any factors involved in plastid gene expression from the ancestral peridinin symbiosis, for example by reinspecting existing transcriptome data (Minge et al. 2010; Burki et al. 2014). Further studies of dinoflagellates that have undergone serial endosymbiosis may provide important insights into the coevolution of plastid genomes and gene expression pathways.
Materials and Methods
Cultures
Karlodinium veneficum RCC2539 (also listed as UIO297) and L. chlorophorum (AC195) were grown in modified k/2 medium, as previously described (Dorrell and Howe 2012), under 50 μE m−2 s−1 continuous light at a controlled temperature of 15 °C. Kryptoperidinium (Glenodinium) foliaceum PCC499 was grown in f/2 medium, under a 30 μE m−2 s−1 12:12 light:dark cycle, at 15–20 °C. To confirm the identity of the Karl. veneficum culture, molecular barcode sequences were generated by PCR of genomic DNA for multiple loci in the Karl. veneficum plastid genome. These were found to be identical to the previously published Karl. veneficum plastid genome sequence (strain UIO083).
Nucleic Acid Isolation
Nucleic acids were isolated from cultures of each species harvested in early stationary phase (ca. 30–60 days postinoculation). Cells were pelleted by centrifugation and washed in sterile growth medium. For RNA isolation, 50 mg pellets of each culture were resuspended in 1 ml TRIzol reagent (Ambion), and frozen at −80 °C and thawed on ice to lyse the cells. Total cellular RNA was then isolated by phase extraction, DNase treated and cleaned with an RNeasy column (Qiagen) as previously described (Barbrook et al. 2012; Dorrell et al. 2014). Genomic DNA was isolated from cell pellets by phase extraction and cleaned with a DNeasy column as previously described (Barbrook and Howe 2000; Nash et al. 2007).
The concentration of each nucleic acid obtained was quantified using a nanodrop spectrophotometer. RNA integrity was confirmed by electrophoresis of 1 μg of each sample in an RNase-free 1% agarose gel containing 0.003% volumes of ethidium bromide. To determine whether any sample contained residual DNA contamination, each RNA sample was used as the direct substrate for a PCR using internal primers against the psbA gene of each sequence. Only samples for which negative results were observed in the initial PCR, and in the product of a reamplification PCR using the initial product as a PCR template, were used for further experimentation.
RT-PCR and Sequencing
Reverse transcription was performed using Superscript III (Life Technologies), as previously described (Dorrell et al. 2014). cDNA was synthesized either with an oligo-d(A) primer, to generate products from polyuridylylated transcripts as previously described (Barbrook et al. 2012) or with internal primers specific to a particular plastid gene. PCR was performed with GoTaq flexi polymerase (Promega) as previously described (Dorrell and Howe 2012). PCR primers used are shown in supplementary table S1, Supplementary Material online. Circular RT-PCR of Karl. veneficum transcripts and thermal asymmetric interlaced PCR of dnaK genetic elements were performed as previously described (Liu et al. 1995; Dorrell and Howe 2012).
PCR products were visualized by electrophoresis in a 1% agarose-TBE gel containing ethidium bromide. Products were directly purified using a QIAquick column elution kit (Qiagen). Where multiple bands were detectable, individual products were separated by electrophoresis, cut out of the agarose gel, and purified as before. Products were sequenced using an Applied Biosystems 3730xl DNA Analyzer. The sequences of three polyuridylylated transcript sequences (psaC, psbI, and psbK) and one internal transcript sequence (ORF4) that were too short to be uploaded to GenBank are listed in supplementary table S2, Supplementary Material online.
Sequence Analysis
Potential recombination events associated with the Karl. veneficum plastid were identified by comparison of the complete plastid genome sequence with the complete plastid genomes of the free-living haptophytes E. huxleyi, P. globosa, Pavlova lutheri, and the partial plastid genome of the uncultured prymnesiophyte C19487 (Puerta et al. 2005; Baurain et al. 2010; Cuvelier et al. 2010).
Poly(U) sites were identified by aligning the sequences of the oligo-d(A) RT-PCR products against the published Karl. veneficum plastid genome sequence (Gabrielsen et al. 2011) using GENEious (www.geneious.com). To identify motifs that might be associated with poly(U) sites, alignments were constructed of the 3′-UTR of each polyuridylylated transcript and of the first 100 bp downstream of the poly(U) site (supplementary table S2, Supplementary Material online). As a negative control, sequence alignments were constructed using the first 100 bp of the 3′-UTR sequence of each gene found not to have a poly(U) site (supplementary table S2, Supplementary Material online). The presence of primary sequence motifs that might be associated with poly(U) sites was investigated by reciprocal BLASTn searches in each alignment, and conserved RNA secondary structures were searched for using the WAR server (http://genome.ku.dk/resources/war/, last accessed June 18, 2014) (Torarinsson and Lindgreen 2008). The relative GC and purine/pyrimidine contents of each sequence were quantified using GENEious, and the minimum Gibbs free energy of folding of each sequence was calculated using the mFold server (http://mfold.rna.albany.edu, last accessed June 18, 2014) (Zuker 2003).
Editing Analysis
Sequence editing was quantified for each gene by GENEious alignments of transcript and genomic sequences. The predicted effect of editing on protein sequence was determined by in silico translation. To determine the effect of transcript editing on protein sequence conservation between Karl. veneficum and haptophyte orthologs, conceptual translation sequences of the transcript and genomic sequence of each gene in the Karl. veneficum were aligned to plastid protein sequences from the haptophytes E. huxleyi and P. globosa using BLAST (Puerta et al. 2005). For each alignment, the number of residues conserved between the Karl. veneficum and haptophyte protein sequences were recorded. Identical amino acids between the two species at any position were scored as a complete match, and positives were scored as a 50% match.
To determine whether editing sites were clustered within certain regions of Karl. veneficum plastid genes, transcript sequences covering the entire CDS of the psaA and tufA genes were obtained by RT-PCR and aligned to the corresponding genomic sequences. Editing sites were identified in each alignment, and scored over a 60-bp sliding sequence window, and regions with elevated frequencies of editing relative to the entire CDS were identified by a binomial test. Sequence conservation between the Karl. veneficum and E. huxleyi protein sequences was scored over each window using BLAST alignment, as before. The total number of matching positions were summed over each 60-bp sliding window, and the Pearson correlation coefficients between the degree of sequence conservation and proportion of edited residues over each gene were calculated.
Supplementary Material
Supplementary figures S1–S8 and tables S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by ASSEMBLE (EFETPIDC), and the BBSRC (BB/F017464/1 to R.G.D.). The authors thank two anonymous referees for their helpful comments on the manuscript. This work was supported by ASSEMBLE (EFETPIDC), the Biotechnology and Biological Sciences Research Council (BB/F017464/1 to R.G.D.) and UCAM OpenAccess (OA-1045).
References
- Asakura Y, Bayraktar OA, Barkan A. Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA. 2008;14:2319–2332. doi: 10.1261/rna.1223708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachvaroff TR, Concepcion GT, Rogers CR, Herman EM, Delwiche CF. Dinoflagellate expressed indicate massive transfer to the nuclear genome sequence tag data of chloroplast genes. Protist. 2004;155:65–78. doi: 10.1078/1434461000165. [DOI] [PubMed] [Google Scholar]
- Barbrook AC, Dorrell RG, Burrows J, Plenderleith LJ, Nisbet RER, Howe CJ. Polyuridylylation and processing of transcripts from multiple gene minicircles in chloroplasts of the dinoflagellate Amphidinium carterae. Plant Mol Biol. 2012;79:347–357. doi: 10.1007/s11103-012-9916-z. [DOI] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ. Minicircular plastid DNA in the dinoflagellate Amphidinium operculatum. Mol Gen Genet. 2000;263:152–158. doi: 10.1007/s004380050042. [DOI] [PubMed] [Google Scholar]
- Baurain D, Brinkmann H, Petersen J, Rodriguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 2010;27:1698–1709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- Bergholtz T, Daugbjerg N, Moestrup O, Fernandez-Tejedor M. On the identity of Karlodinium veneficum and description of Karlodinium armiger sp nov (Dinophyceae), based on light and electron microscopy, nuclear-encoded LSU rDNA, and pigment composition. J Phycol. 2006;42:170–193. [Google Scholar]
- Burki F, Imanian B, Hehenberger E, Hirakawa Y, Maruyama S, Keeling PJ. Endosymbiotic gene transfer in tertiary plastid-containing dinoflagellates. Eukaryot Cell. 2014;13:246–255. doi: 10.1128/EC.00299-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvelier ML, Allen AE, Monier A, McCrow JP, Messie M, Tringe SG, Woyke T, Welsh RM, Ishoey T, Lee JH, et al. Targeted metagenomics and ecology of globally important uncultured eukaryotic phytoplankton. Proc Natl Acad Sci U S A. 2010;107:14679–14684. doi: 10.1073/pnas.1001665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Green BR. Substitutional editing of Heterocapsa triquetra chloroplast transcripts and a folding model for its divergent chloroplast 16S rRNA. Gene. 2009;442:73–80. doi: 10.1016/j.gene.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Dorrell RG, Drew J, Nisbet RE, Howe CJ. Evolution of chloroplast transcript processing in Plasmodium and its chromerid algal relatives. PLoS Genet. 2014;10:e1004008. doi: 10.1371/journal.pgen.1004008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell RG, Howe CJ. Functional remodeling of RNA processing in replacement chloroplasts by pathways retained from their predecessors. Proc Natl Acad Sci U S A. 2012;109:18879–18884. doi: 10.1073/pnas.1212270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelund M, Minge MA, Gabrielsen TM, Nederbragt AJ, Shalchian-Tabrizi K, Otis C, Turmel M, Lemieux C, Jakobsen KS. Genome fragmentation is not confined to the peridinin plastid in dinoflagellates. PLoS One. 2012;7:e38809. doi: 10.1371/journal.pone.0038809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Small I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011;191:37–47. doi: 10.1111/j.1469-8137.2011.03746.x. [DOI] [PubMed] [Google Scholar]
- Gabrielsen TM, Minge MA, Espelund M, Tooming-Klunderud A, Patil V, Nederbragt AJ, Otis C, Turmel M, Shalchian-Tabrizi K, Lemieux C, et al. Genome evolution of a tertiary dinoflagellate plastid. PLoS One. 2011;6:e19132. doi: 10.1371/journal.pone.0019132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66:34–44. doi: 10.1111/j.1365-313X.2011.04541.x. [DOI] [PubMed] [Google Scholar]
- Hayes ML, Giang K, Mulligan RM. Molecular evolution of pentatricopeptide repeat genes reveals truncation in species lacking an editing target and structural domains under distinct selective pressures. BMC Evol Biol. 2012;12:13. doi: 10.1186/1471-2148-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe CJ, Nisbet RER, Barbrook AC. The remarkable chloroplast genome of dinoflagellates. J Exp Bot. 2008;59:1035–1045. doi: 10.1093/jxb/erm292. [DOI] [PubMed] [Google Scholar]
- Iida S, Kobiyama A, Ogata T, Murakami A. Identification of transcribed and persistent variants of the psbA gene carried by plastid minicircles in a dinoflagellate. Curr Genet. 2009;55:583–591. doi: 10.1007/s00294-009-0271-9. [DOI] [PubMed] [Google Scholar]
- Imanian B, Pombert JF, Keeling PJ. The complete plastid genomes of the two ‘dinotoms’ Durinskia baltica and Kryptoperidinium foliaceum. PLoS One. 2010;5:e10711. doi: 10.1371/journal.pone.0010711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, Gornik SG, Waller RF. A tertiary plastid gains RNA editing in its new host. Mol Biol Evol. 2013;30:788–792. doi: 10.1093/molbev/mss270. [DOI] [PubMed] [Google Scholar]
- Janouskovec J, Horák A, Oborník M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A. 2010;107:10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouskovec J, Sobotka R, Lai DH, Flegontov P, Koník P, Komenda J, Ali S, Prásil O, Pain A, Oborník M, et al. Split photosystem protein, linear-mapping topology, and growth of structural complexity in the plastid genome of Chromera velia. Mol Biol Evol. 2013;30:2447–2462. doi: 10.1093/molbev/mst144. [DOI] [PubMed] [Google Scholar]
- Lange H, Sement FM, Canaday J, Gagliardi D. Polyadenylation-assisted RNA degradation processes in plants. Trends Plant Sci. 2009;14:497–504. doi: 10.1016/j.tplants.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Larson EM, Obrien CM, Zhu GH, Spreitzer RJ, Portis AR. Specificity for activase is changed by a Pro-89 to Arg substitution in the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. J Biol Chem. 1997;272:17033–17037. doi: 10.1074/jbc.272.27.17033. [DOI] [PubMed] [Google Scholar]
- Li CH, Salvucci ME, Portis AR. Two residues of rubisco activase involved in recognition of the rubisco substrate. J Biol Chem. 2005;280:24864–24869. doi: 10.1074/jbc.M503547200. [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Shinozaki F, Chikuni T, Yabuki A, Takishita K, Kawachi M, Nakayama T, Inouye I, Hashimoto T, Inagaki Y. Green-colored plastids in the dinoflagellate genus Lepidodinium are of core chlorophyte origin. Protist. 2011;162:268–276. doi: 10.1016/j.protis.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Minge MA, Shalchian-Tabrizi K, Torresen OK, Takishita K, Probert I, Inagaki Y, Klaveness D, Jakobsen KS. A phylogenetic mosaic plastid proteome and unusual plastid-targeting signals in the green-colored dinoflagellate Lepidodinium chlorophorum. BMC Evol Biol. 2010;10:191. doi: 10.1186/1471-2148-10-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungpakdee S, Shinzato C, Takeuchi T, Kawashima T, Koyanagi R, Hisata K, Tanaka M, Goto H, Fujie M, Lin S, et al. Massive gene transfer and extensive RNA editing of a symbiotic dinoflagellate plastid genome. Genom Biol Evol. 2014;6:1408–1422. doi: 10.1093/gbe/evu109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash EA, Barbrook AC, Edwards-Stuart RK, Bernhardt K, Howe CJ, Nisbet RER. Organization of the mitochondrial genome in the dinoflagellate Amphidinium carterae. Mol Biol Evol. 2007;24:1528–1536. doi: 10.1093/molbev/msm074. [DOI] [PubMed] [Google Scholar]
- Nelson MJ, Dang YK, Filek E, Zhang ZD, Yu VWC, Ishida K, Green BR. Identification and transcription of transfer RNA genes in dinoflagellate plastid minicircles. Gene. 2007;392:291–298. doi: 10.1016/j.gene.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Puerta MVS, Bachvaroff TR, Delwiche CF. The complete plastid genome sequence of the haptophyte Emiliania huxleyi: a comparison to other plastid genomes. DNA Res. 2005;12:151–156. doi: 10.1093/dnares/12.2.151. [DOI] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Skanseng M, Ronquist F, Klaveness D, Bachvaroff TR, Delwiche CF, Botnen A, Tengs T, Jakobsen KS. Heterotachy processes in rhodophyte-derived secondhand plastid genes: implications for addressing the origin and evolution of dinoflagellate plastids. Mol Biol Evol. 2006;23:1504–1515. doi: 10.1093/molbev/msl011. [DOI] [PubMed] [Google Scholar]
- Takano Y, Hansen G, Fujita D, Horiguchi T. Serial replacement of diatom endosymbionts in two freshwater dinoflagenates, Peridiniopsis spp. (Peridiniales, Dinophyceae) Phycologia. 2008;47:41–53. [Google Scholar]
- Takishita K, Kawachi M, Noel M-H, Matsumoto T, Kakizoe N, Watanabe MM, Inouye I, Ishida K-I, Hashimoto T, Inagaki Y. Origins of plastids and glyceraldehyde-3-phosphate dehydrogenase genes in the green-colored dinoflagellate Lepidodinium chlorophorum. Gene. 2008;410:26–36. doi: 10.1016/j.gene.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Takishita K, Nakano K, Uchida A. Preliminary phylogenetic analysis of plastid-encoded genes from an anomalously pigmented dinoflagellate Gymnodinium mikimotoi (Gymnodiniales, Dinophyta) Phycol Res. 1999;47:257–262. [Google Scholar]
- Torarinsson E, Lindgreen S. WAR: Webserver for aligning structural RNAs. Nucl Acids Res. 2008;36:79–84. doi: 10.1093/nar/gkn275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Morse D. Rampant polyuridylylation of plastid gene transcripts in the dinoflagellate Lingulodinium. Nucl Acids Res. 2006;34:613–619. doi: 10.1093/nar/gkj438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauner S, Greilinger D, Laatsch T, Kowallik KV, Maier UG. Substitutional editing of transcripts from genes of cyanobacterial origin in the dinoflagellate Ceratium horridum. FEBS Lett. 2004;577:535–538. doi: 10.1016/j.febslet.2004.10.060. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Green BR, Cavalier-Smith T. Single gene circles in dinoflagellate chloroplast genomes. Nature. 1999;400:155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.