Figure 3.
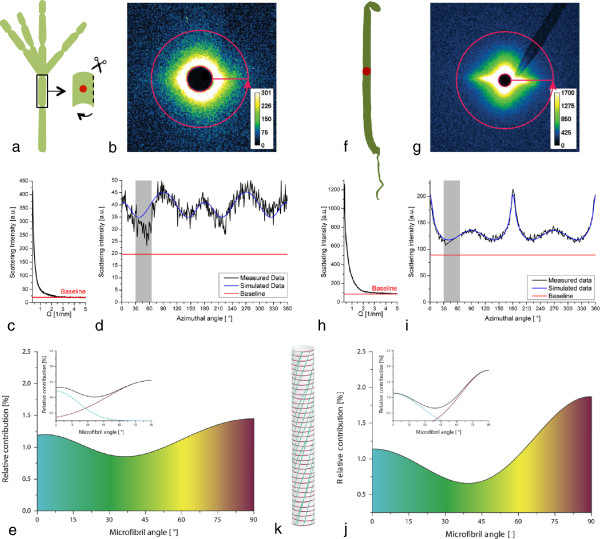
SAXS analysis.a-e: Chara corallina. a) One cell was cut open and a single cell wall was analysed. f-j: Primary cell walls of Arabidopsis thaliana. a and f) 6 day old dark grown hypocotyl is analysed. The beam diameter is close to the hypocotyl diameter. b and g) 2D scattering image: 360° azimuthal integration is marked. The scattering signal originates from electron density contrast between the cellulose microfibrils and water. c and h) The radial scattering profile is used to assign the background of the measurement. d and i) Integrated scattering signal (measured data) can be fitted by microfibril angle simulation (simulated data). The area beneath the baseline as well as the data range influenced by the glass capillary on which the beamstop was mounted (grey area) are not considered in the simulation. e and j) Relative contribution of each microfibril angle to the simulated scattering signal. The colour gradient represents the microfibril orientations from 0° (longitudinal towards the cell axis, indicated in blue) to 90° (transverse to the longitudinal axis of the cell, indicated in red). The insets show the two underlying distributions. At intermediate microfibril angles there is an overlap in which the two distributions cannot be distinguished. k) Schematic of a possible arrangement of the microfibrils in a cell with the colour scheme used in e and j.
