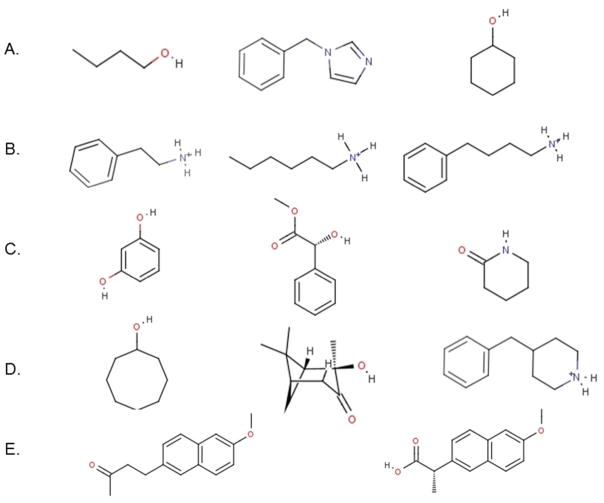
Figure 2.
Chemical structures of 14 representative guests taken from the set of 57 guests investigated (see Supplementary Material for a complete list). Each row corresponds to a class of guest discussed in the text: (A) small guests with single functionalities well described by the model (this class comprises 17 members), (B) protonated amines (22 members) and (C) phenols, esters, and amides (6 members) with functionalities described less well by the model, (D) large and nominally rigid guests, and strong binders (10 members) believed to undergo complex reorganization upon binding, and (E) nabumetone and naproxen, which are correctly predicted as some of the strongest binders by the model.