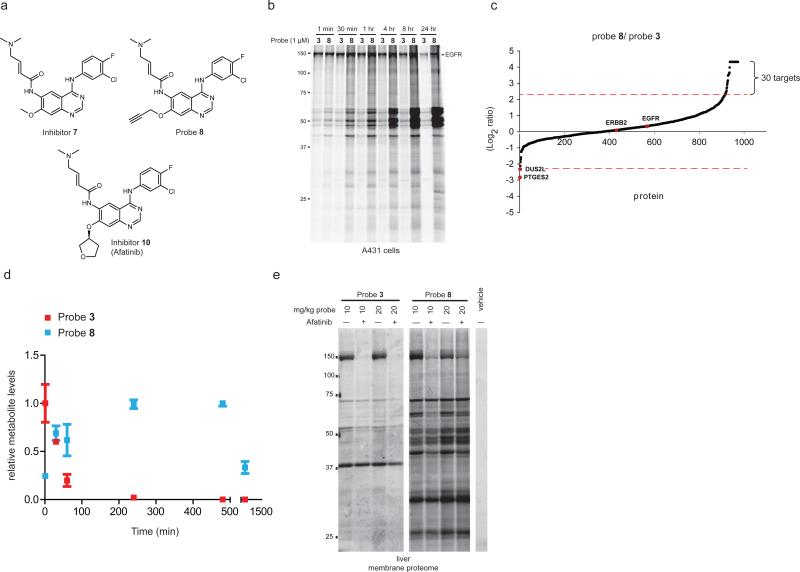
Figure 5.
The in situ proteomic reactivity of covalent kinase inhibitors is altered by modification of the Michael acceptor. (a) Structures of dimethylaminomethyl (DMAM)-modified agents [inhibitor 7, probe 8, and afatinib (10)]. (b) Gel-based ABPP showing markedly different in situ reactivity profiles for probes 3 and 8 (1 μM) in A431 cells treated for the indicated time. (c) SILAC ratios for probe targets from a probe-probe comparison study performed in duplicate, where cancer cells were treated with probe 3 (1 uM, 4 hr) or 8 (1 μM, 4 hr). Results are plotted as Log2 of the median SILAC ratios (probe 8/probe 3). Proteins exhibiting preferential labeling with probe 8 or 3 extend above and below the x-axis, respectively. Proteins with SILAC ratios ≥ 5 are designated as probe-selective targets. (d) Time course measurement of probe 3 (red) and 8 (green) concentrations in A431 cells. Data represent relative amounts of each compound normalized to the highest measured cellular concentration across the time course (set to an arbitrary value of 1.0) and are reported as average values + standard errors for three biological replicates. (e) Gel-based ABPP of liver membrane proteomes from mice treated with probes 3 or 8 (10 or 20 mg/kg, 1 hr). Pre-treatment with afatinib (20 mg/kg, 1 hr) blocked probe-labeling of an ~150 kDa protein interpreted to represent EGFR. For (b, d), gel-based ABPP experiments were performed in duplicate or triplicate with consistent results.