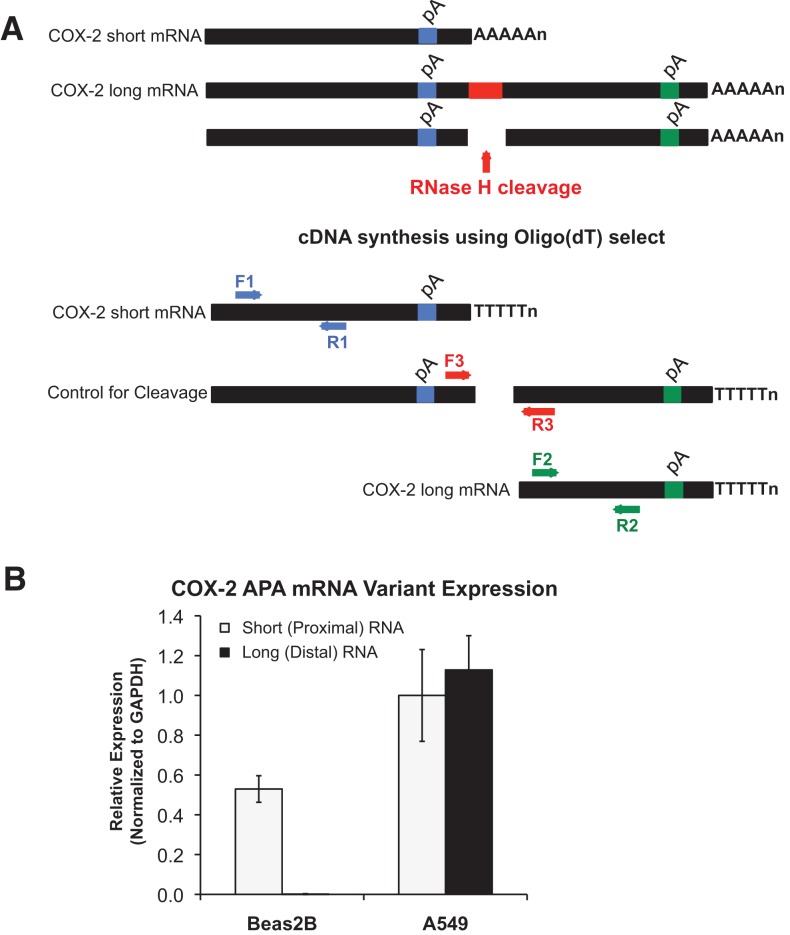
FIGURE 2.
Alternative polyadenylation of COX-2. (A) Schematic of COX-2 3′ UTR depicting the relative location of PCR primer sets that are COX-2 mRNA variant specific. Since Oligo(dT) selection was used in the cDNA synthesis reaction, the F1:R1 primer set will specifically amplify the COX-2 short polyadenylated mRNA; the F2:R2 set will specifically amplify the COX-2 long mRNA; the F3:R3 set will ensure that RNase H cleavage reaction took place. Use of the F3:R3 primer set resulted in no amplified PCR product, indicating that RNase H cleavage was complete (data not shown). (B) Quantification of COX-2 alternative polyadenylation (APA) mRNA variant expression as measured by RHAPA analysis revealed expression of both COX-2 mRNA transcripts (short and long) in A549 cells as compared with Beas2B cells. COX-2 expression was normalized to GAPDH mRNA; n = 4.