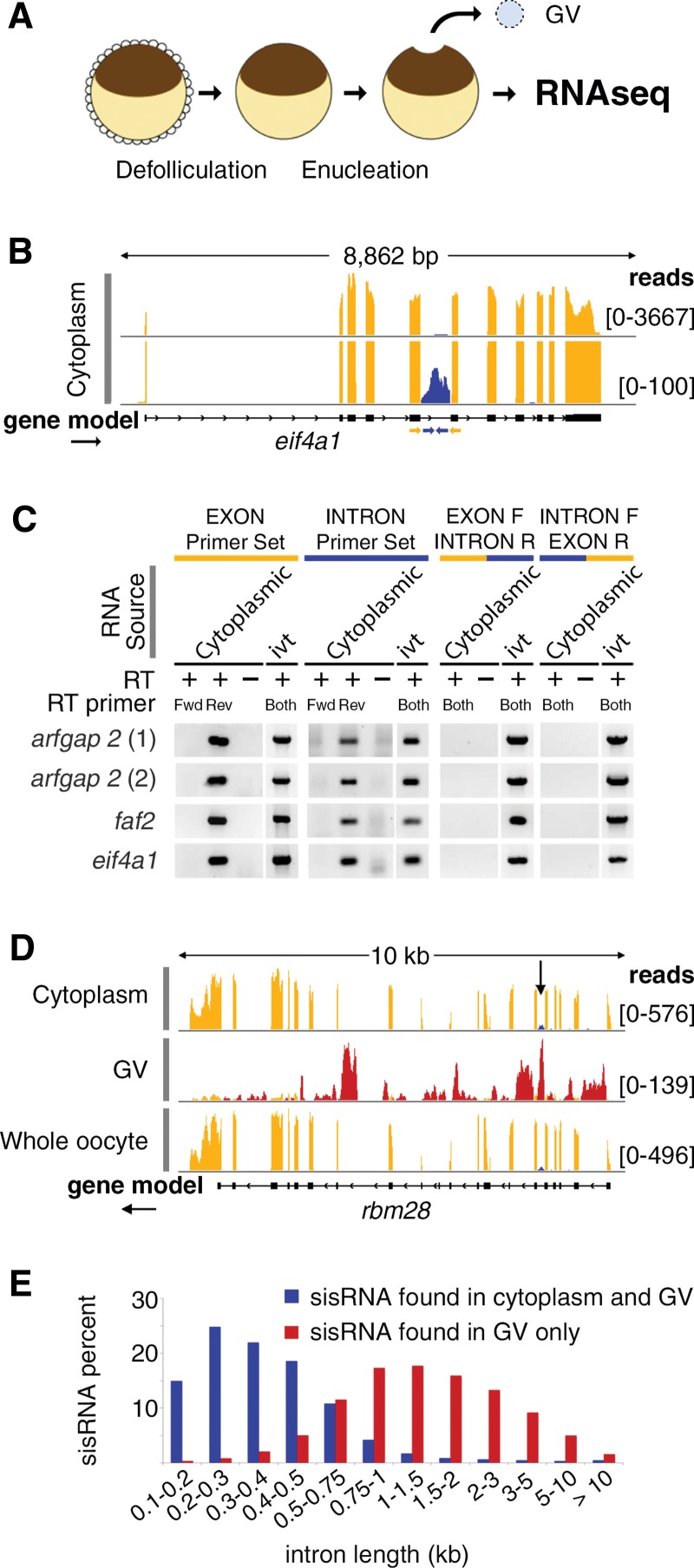
FIGURE 1.
Characterization of cytoplasmic sisRNAs. (A) The oocyte nucleus or germinal vesicle (GV) can be removed from a defolliculated oocyte within seconds to provide a sample of cytoplasm uncontaminated with nuclear RNA. (B) IGV browser view of a typical gene bearing a cytoplasmic sisRNA. Cytoplasmic intronic reads (blue) occur in one intron of the eif4a1 gene (yellow exonic reads). The lower track (track height 100 reads) shows that the intronic reads do not cross the intron–exon borders. The yellow and blue arrows represent exonic and intronic primers used for RT–PCR. (C) RT–PCR results showing that cytoplasmic sisRNAs are derived from the sense strand and do not cross the intron–exon borders. (ivt) In vitro transcribed RNA, used as a control template. (D) Comparison of cytoplasmic, nuclear (GV), and whole oocyte RNAs from the rbm28 gene. Cytoplasmic and whole oocyte RNAs are essentially identical (arrow points to cytoplasmic sisRNA, blue reads). Nuclear sisRNAs (red reads in GV track) are not detectable in the whole oocyte sample. (E) Distribution of sisRNAs relative to intron length. The majority of cytoplasmic sisRNAs (blue) derive from shorter introns, whereas nuclear-specific sisRNAs (red) come from a wide range of longer introns.