Figure 2. The epigenetic marks that define the transcriptional state of an ES.
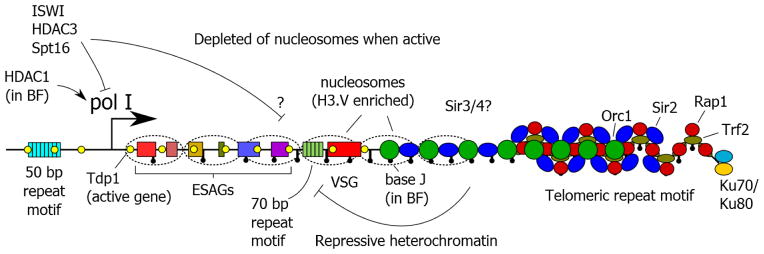
A repressive chromatin structure is formed by TbTRF2 and TbRAP1 (which may recruit Sir2) as well as TbORC1, propagating to sub-telomeric regions. It is not known whether other proteins fulfil the roles of yeast Sir3 and Sir4, for which orthologues are absent in T. brucei. Base J is present at an increasing density towards the telomere termini, and is required for ES silencing. Nucleosomes present on a silent ES are enriched for the transcriptional terminating variant H3.V, and are depleted on an active ES. The HMG box protein TbTDP1 is present on the active ES, and is associated with chromatin decondensation. The histone deacetylase TbHDAC3 and the chromatin remodeller TbISWI is required for efficient ES silencing, and TbHDAC1 is required for activated expression.
