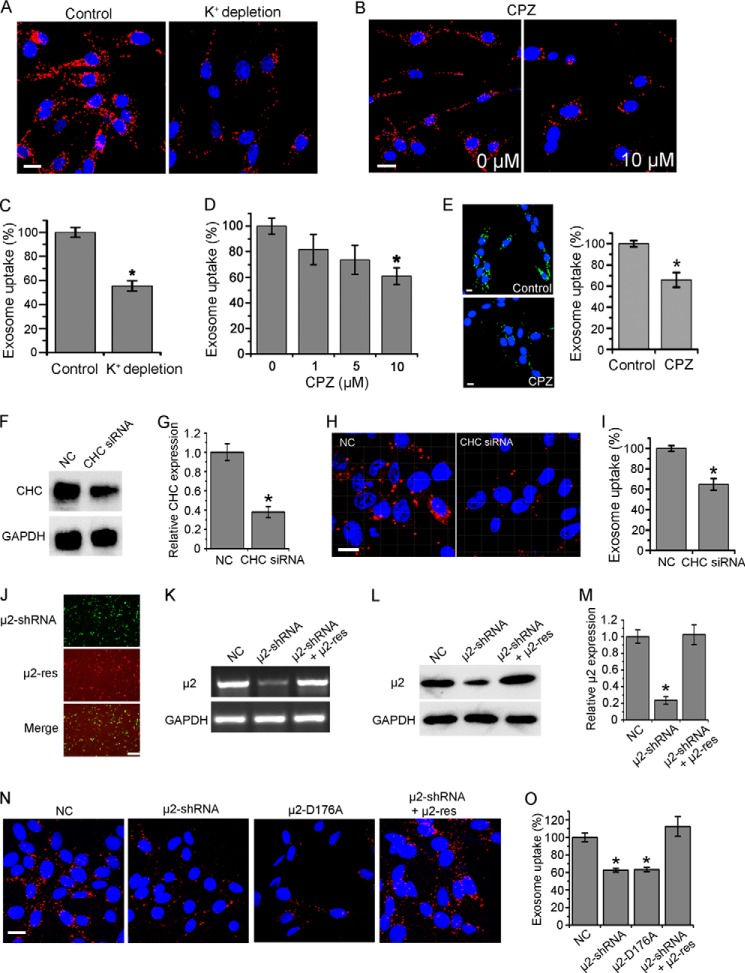
FIGURE 2.
Role of clathrin-mediated endocytosis in exosome uptake. A, confocal images of PC12 cells incubated with DiD-labeled exosomes at 37 °C for 3 h in K+ depletion buffer with (as control) or without KCl. B, confocal images of PC12 cells pretreated with CPZ or not for 30 min, and then incubated with exosomes at 37 °C for 3 h. C and D, exosome uptake under various treatments and control was quantified by determining the fluorescence intensity. E, confocal images of PC12 cells pretreated with 10 μm CPZ or not for 30 min, and then incubated with CFSE-labeled exosomes at 37 °C for 3 h. Exosome uptake was quantified by determining the fluorescence intensity. All scale bars above, 15 μm. F, Western blots of CHC after 48 h of treatment of corresponding siRNA. G, CHC expression level was quantified by determining the gray value. H, confocal images of cells pretreated with siRNA against CHC or NC for 48 h, and then incubated with exosomes at 37 °C for 3 h. Scale bar, 20 μm. I, exosome uptake was quantified by determining the fluorescence intensity. J, fluorescence images of PC12 cells transfected with μ2-shRNA (GFP tagged), μ2-res (mCherry tagged), and merged together. Scale bar, 50 μm. PCR (K) and Western blot (L) results of μ2 48 h after transfection of NC, μ2-shRNA, or μ2-shRNA and μ2-res together. M, μ2 expression level was quantified by determining the gray value. N, confocal images of cells 48 h after transfection of NC, μ2-shRNA, μ2-D176A, or μ2-shRNA and μ2-res together, and then incubated with exosomes at 37 °C for 3 h. Scale bar, 20 μm. O, exosome uptake was quantified by determining the fluorescence intensity. In all the confocal images, red refers to DiD-labeled exosomes, and blue indicates nuclei. All the values are normalized to the control. For all the quantification plots, mean ± S.D. of three independent experiments is shown. Values significantly different (p < 0.05) from control are marked with asterisks.