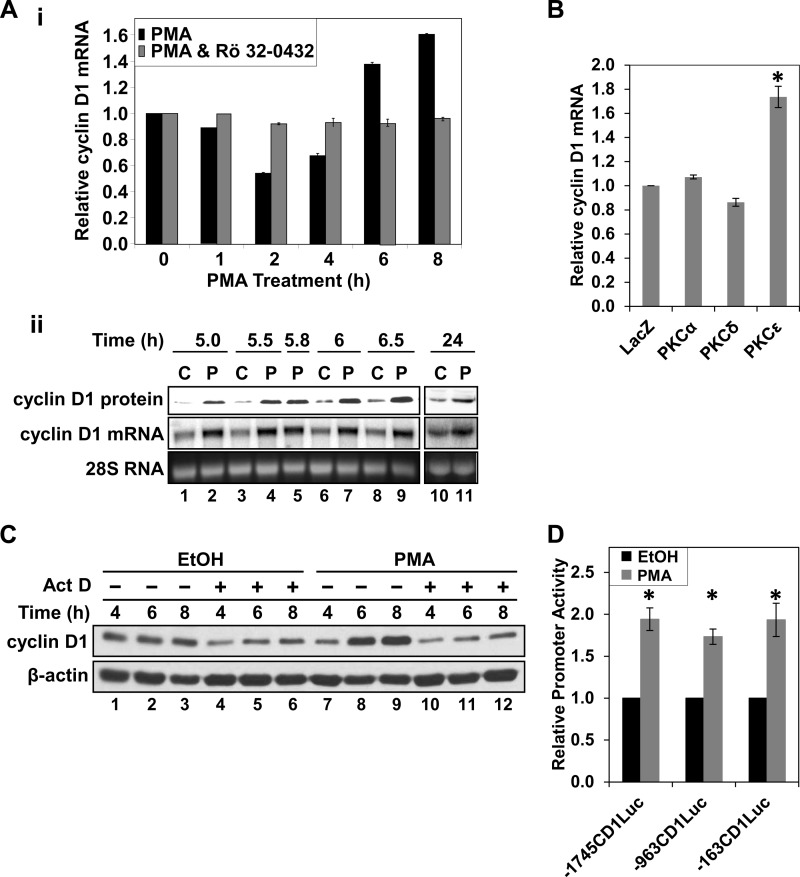
FIGURE 5.
PKCϵ induces cyclin D1 mRNA in IEC-18 cells through activation of the proximal cyclin D1 promoter. A, effects of PMA on cyclin D1 mRNA expression. i, cells were treated with PMA in the presence or absence of the PKC inhibitor Rö32-0432 (5 μm) for the indicated times, and cyclin D1 mRNA levels were analyzed by quantitative real-time PCR. Data were normalized to 18 S rRNA and expressed as -fold change relative to that in cells treated with vehicle (ethanol). Data are averages ± S.E. (error bars) of three independent experiments. ii, cells were treated with PMA or vehicle for the indicated times and analyzed for cyclin D1 mRNA and protein levels by Northern and Western blotting, respectively. B, overexpression of PKCϵ induces cyclin D1 mRNA expression. Cells were transduced with adenoviral vectors (multiplicity of infection 20) expressing either LacZ, PKCα, PKCδ, or PKCϵ, and expression of cyclin D1 mRNA was assessed and normalized to 18 S rRNA as in A. Data are expressed relative to levels in LacZ-transduced cells and are averages ± S.E. of more than three independent experiments. *, statistically different from LacZ-transduced cells (p < 0.05). C, hyperinduction of cyclin D1 requires new transcription. Cells were pretreated with 5 μg/ml actinomycin D (Act D) or vehicle (DMSO) for 30 min prior to the addition of PMA or vehicle (EtOH) for the indicated times. Cyclin D1 and β-actin protein levels were analyzed by Western blotting. Data are representative of more than three independent experiments. D, PMA activates the proximal (−163 bp) cyclin D1 promoter. Cells were transfected with promoter-luciferase constructs representing the proximal 1745 (−1745CD1Luc), 963 (−936CD1Luc), or 163 bp (−163CD1Luc) of the cyclin D1 promoter and treated with PMA or vehicle for 22 h. Promoter activity was assessed from luciferase expression and is expressed relative to that in control (vehicle-treated) cells for each construct. Data are averages ± S.E. of three independent experiments. *, statistically different from vehicle (EtOH)-treated control cells (p < 0.05).