FIGURE 6.
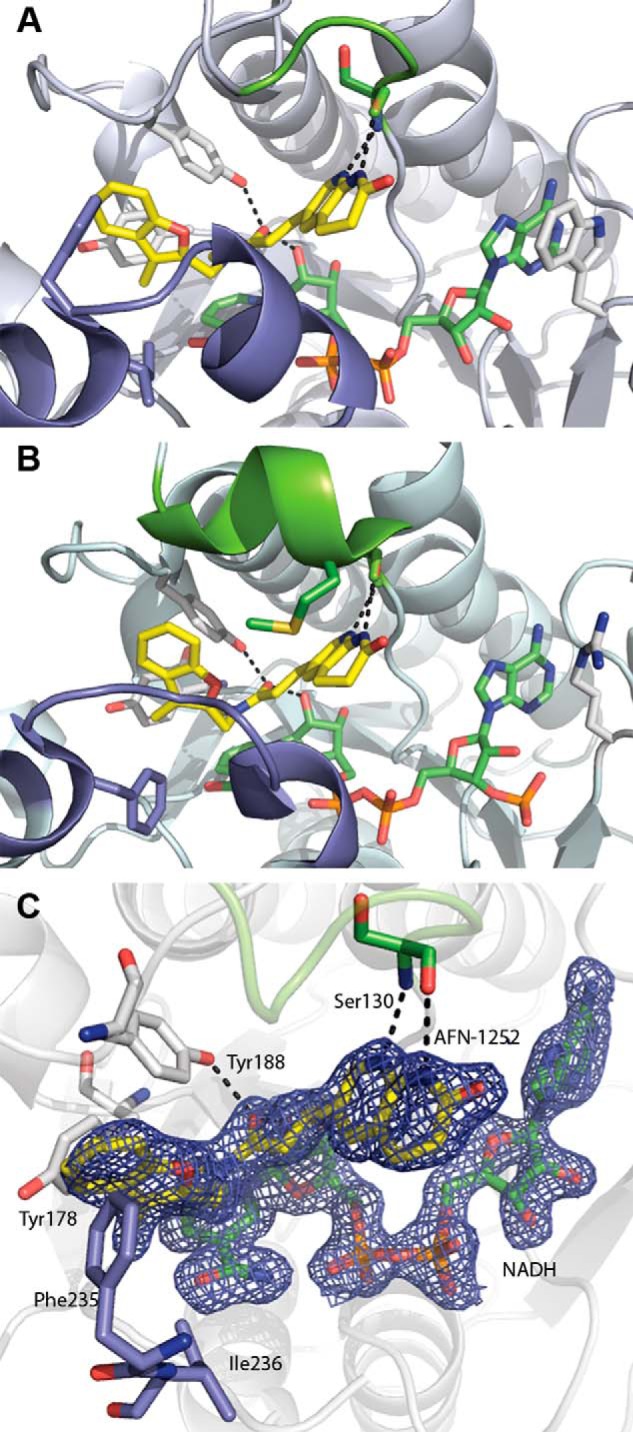
FabI·NADH·AFN ternary complex structure. A, CtFabI·NADH·AFN-152 ternary complex (Protein Data Bank accession code 4Q9N). B, SaFabI·NADPH·AFN-1252 ternary complex (Protein Data Bank accession code 4FS3). The color schemes are the same in A and B. AFN-1252 is shown in yellow, and NADPH/NADH is shown in green. The flexible, substrate-binding flipping loop is colored slate, and the poorly conserved lid loop is colored chartreuse. Black dashes show the conserved hydrogen bonding network for AFN-1252. In SaFabI, the substrate lid adopts a more ordered, helical structure and orients Met-99 into the active site toward the inhibitor. In contrast, in CtFabI, the active site is more open, due to the less structured conformation of the lid. C, active site of the CtFabI·NADH·AFN-1252 ternary complex. AFN-1252 binds within a hydrophobic pocket composed of Tyr-178, Phe-235, and Ile-236, with Phe-235 positioned for π-interactions with the 3-methylbenzofuran of AFN-1252. The 2′-hydroxyl of NADH and the hydroxyl of Tyr-188 make hydrogen bonds with the carbonyl of the linking cis-amide, and the Ser-130 peptide backbone carbonyl and amide make hydrogen bonds with the pyridyl nitrogen and the N-acyl hydrogen of the oxotetrahydronaphthyridine of AFN-1252. σA-weighted 2Fo − Fc electron density is shown for AFN-1252 and NADH contoured at 1.5 σ.
