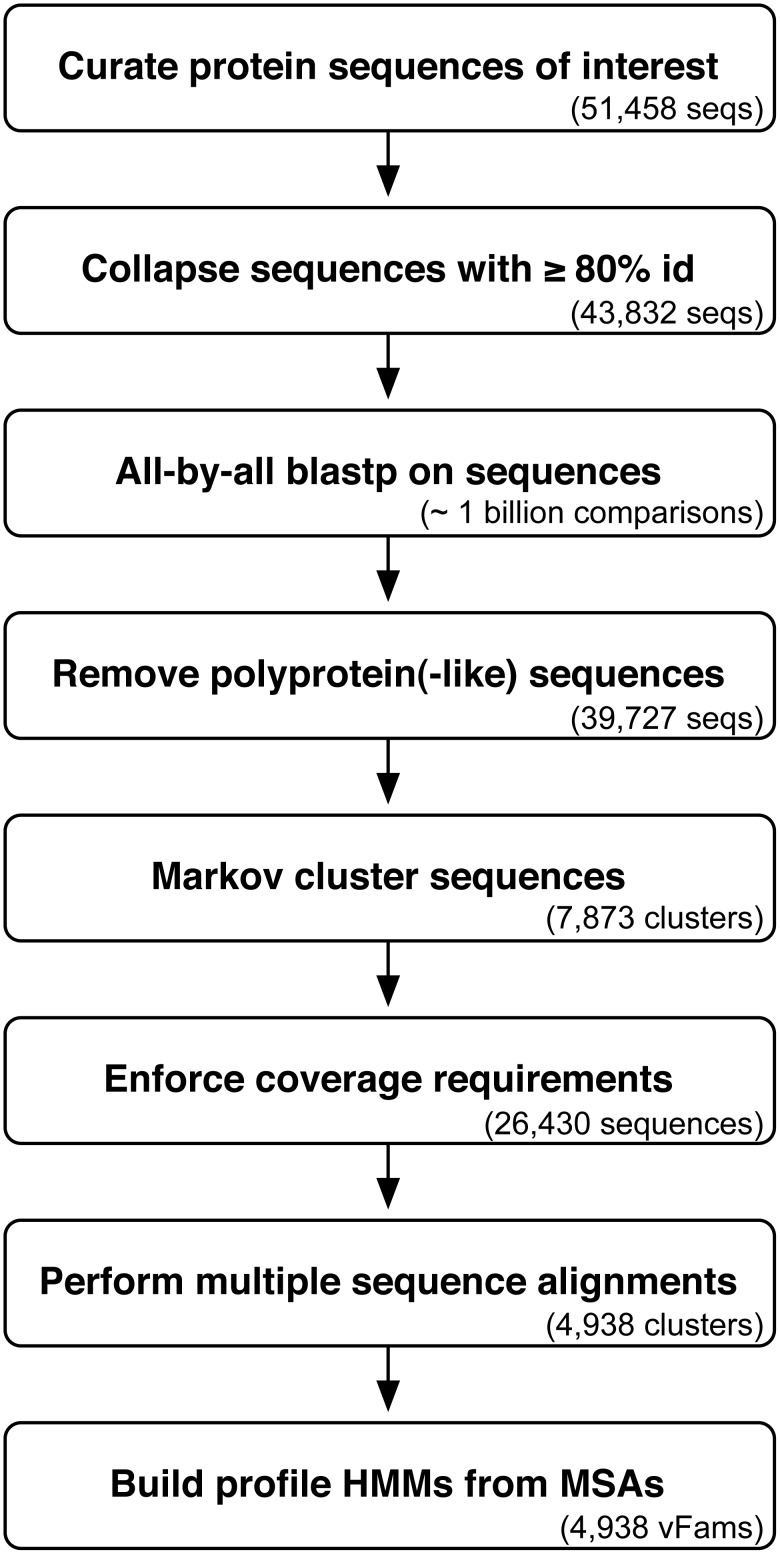
Figure 1. The pipeline for building profile HMMs from a set of curated viral protein sequences.
An initial set of protein sequences of interest is curated and reduced by collapsing high-identity sequences. The similarity between all pairs of remaining sequences is calculated using BLAST. Using the BLAST results, polyprotein sequences are inferred and removed. The Markov Clustering algorithm groups the remaining sequences into families. Sequences with extreme lengths are removed before multiple sequence alignments are generated for each family. Multiple sequence alignments are used to train profile HMMs. Statistics for each step in the generation of the vFams are in parentheses.