Figure 4. Performance of vFams and BLAST on metagenomic datasets.
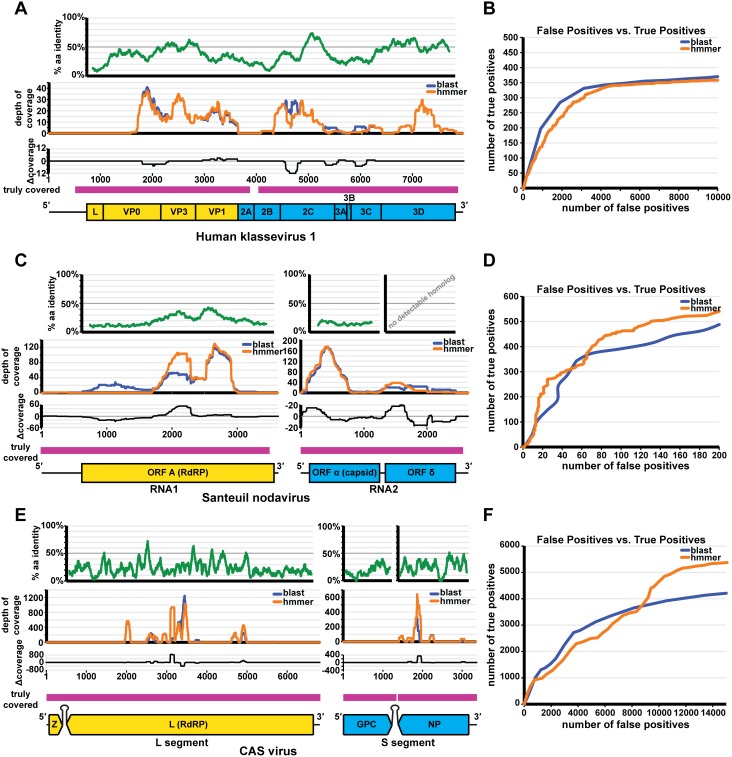
A comparison of BLAST vs. HMMER for the detection of Human klassevirus 1, Santeuil nodavirus, and CAS virus. (A) Percent amino acid identity for 80 aa windows is shown between the Human klassevirus 1 and Aichi virus polyprotein sequences (green); genome coverage of correctly classified viral reads by BLAST and HMMER is shown in blue and orange respectively; the difference in coverage (HMMER coverage−BLAST coverage = Δ coverage) is shown in black; the regions of the genome truly covered in the full dataset are shown in pink; a to-scale genome schematic of Human klassevirus 1 is found below, depicting structural proteins (yellow) and non-structural proteins (blue). (B) The number of true positives vs. the number of false positives for the detection of Human klassevirus 1 is depicted for BLAST (blue) and HMMER (orange). (C) Percent amino acid identity for 84 aa windows is shown between ORF A and ORF α of Santeuil nodavirus and the RdRP and capsid proteins of the Striped Jack Nervous Necrosis virus (green) [no homolog of ORF δ was detected at the time of the discovery] [42]; genome coverage of correctly classified viral reads by BLAST and HMMER is shown in blue and orange respectively; the difference in coverage (HMMER coverage−BLAST coverage = Δ coverage) is shown in black; the regions of the genome truly covered in the full dataset are shown in pink; a to-scale genome schematic of Santeuil nodavirus RNA-1 and RNA-2 is found below, depicting ORF A (yellow), and ORF α and ORF δ (blue). (D) The number of true positives vs. the number of false positives for the detection of Santeuil nodavirus is depicted for BLAST (blue) and HMMER (orange). (E) Percent amino acid identity for 33 aa windows is shown between the L protein, the glycoprotein, and the nucleoprotein of CAS virus and the L protein of Lymphocytic choriomeningitis virus, the glycoprotein of Lloviu virus, and the nucleoprotein of Lymphocytic choriomeningitis virus (green) respectively [no homolog of the Z protein was detected at the time of the discovery] [12]; genome coverage of correctly classified viral reads by BLAST and HMMER is shown in blue and orange respectively; the difference in coverage (HMMER coverage−BLAST coverage = Δ coverage) is shown in black; the regions of the genome truly covered in the full dataset are shown in pink; a genome schematic of the CAS virus L segment and S segment is found below, depicting the Z and L proteins (yellow) and the glycoprotein (GPC) and nucleoprotein (NP) (blue) respectively. (F) The number of true positives vs. the number of false positives for the detection of CAS virus is depicted for BLAST (blue) and HMMER (orange).