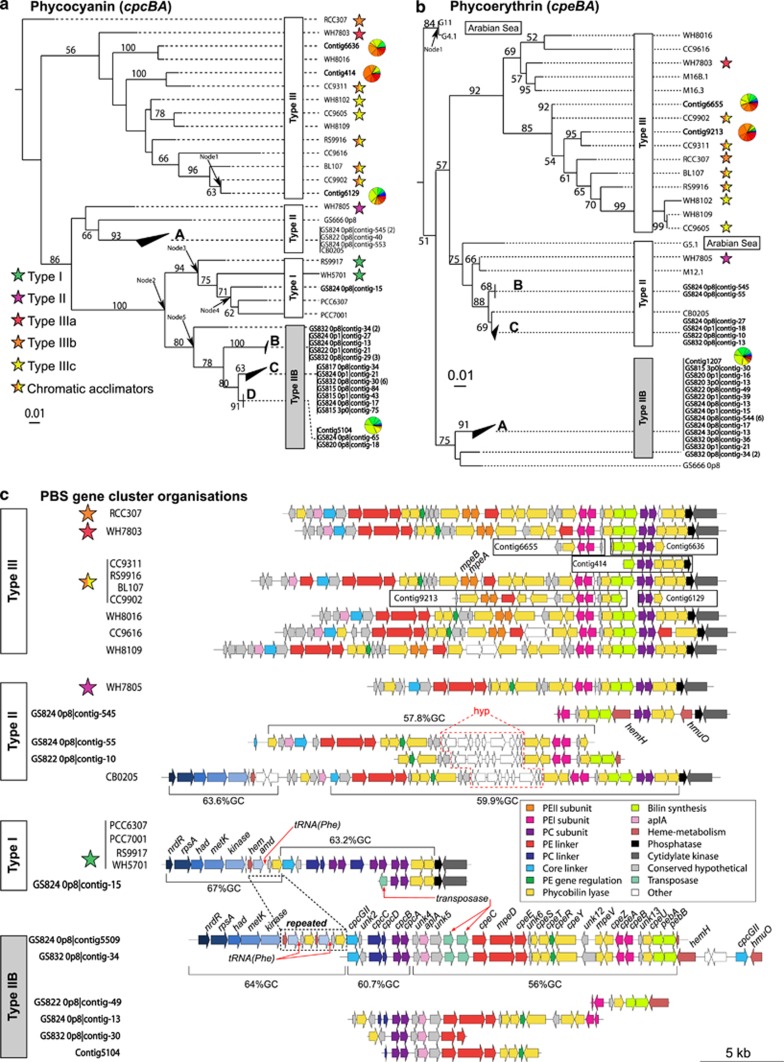
Figure 4.
Phylogeny of phycobiliproteins and structure of the PBS operons. ML phylogenies of concatenated amino-acid alignments of PC (cpcBA) (a) and PE (cpeBA) (b) subunits from reference genomes and assembled contigs from the Baltic Sea metagenomes. The trees were rooted with Gloeobacter violaceus (data not shown). Bootstrap (1000 replicates) support >50% is shown at nodes and the scale bar indicates number of expected substitutions per site. Stars as in Figure 2. For contigs in the 2009 global assembly, pie charts show the read distribution from each site. Numbers in parenthesis next to contigs indicate the number of contigs with identical PC or PE subunits. Arrows show internal nodes introduced by phylogenetic placement of sequence fragments (see Figure 5). (c) PBS operon structures of picocyanobacteria in the data set. Operon schematics are shown for each pigment type and for assembled contigs in the Baltic Sea data set. For contigs that occupy the same position in the PC and PE phylogenies, only the longest contig is shown. The region designated 'hyp' in type II genomes indicates a 4.4-kb stretch of conserved hypothetical genes. amd, predicted amidophosphoribosyltransferase; had, haloacid dehalogenase-like hydrolase; hem, putative heme iron utilization protein; kinase, carbohydrate kinase, FGGY family protein; metK, S-adenosylmethionine synthetase; nrdR, transcriptional regulator NrdR; rpsA, 30S ribosomal protein S1.