Abstract
Background
Rapid activation of innate immune defences upon microbial infection depends on the evolutionary conserved NF-κB dependent signals which deregulation is frequently associated with chronic inflammation and oncogenesis. These signals are tightly regulated by the linkage of different kinds of ubiquitin moieties on proteins that modify either their activity or their stability. To investigate how ubiquitin specific proteases (USPs) orchestrate immune signal regulation, we created and screened a focused RNA interference library on Drosophila NF-κB-like pathways Toll and Imd in cultured S2 cells, and further analysed the function of selected genes in vivo.
Results
We report here that USP2 and USP34/Puf, in addition to the previously described USP36/Scny, prevent inappropriate activation of Imd-dependent immune signal in unchallenged conditions. Moreover, USP34 is also necessary to prevent constitutive activation of the Toll pathway. However, while USP2 also prevents excessive Imd-dependent signalling in vivo, USP34 shows differential requirement depending on NF-κB target genes, in response to fly infection by either Gram-positive or Gram-negative bacteria. We further show that USP2 prevents the constitutive activation of signalling by promoting Imd proteasomal degradation. Indeed, the homeostasis of the Imd scaffolding molecule is tightly regulated by the linkage of lysine 48-linked ubiquitin chains (K48) acting as a tag for its proteasomal degradation. This process is necessary to prevent constitutive activation of Imd pathway in vivo and is inhibited in response to infection. The control of Imd homeostasis by USP2 is associated with the hydrolysis of Imd linked K48-ubiquitin chains and the synergistic binding of USP2 and Imd to the proteasome, as evidenced by both mass-spectrometry analysis of USP2 partners and by co-immunoprecipitation experiments.
Conclusion
Our work identified one known (USP36) and two new (USP2, USP34) ubiquitin specific proteases regulating Imd or Toll dependent immune signalling in Drosophila. It further highlights the ubiquitin dependent control of Imd homeostasis and shows a new activity for USP2 at the proteasome allowing for Imd degradation. This study provides original information for the better understanding of the strong implication of USP2 in pathological processes in humans, including cancerogenesis.
Keywords: Innate immunity, Imd, NF-κB, Proteasome, RNA interference screen, Toll, Ubiquitin specific protease, USP2 (CG14619), USP34 (CG5794), USP36 (CG5505)
Lay abstract
Inflammation is a major player of our innate immune defences which is induced within minutes following infection by microorganisms. Its activation mainly depends on intracellular signals inducing the secretion of pro-inflammatory cytokines and anti-microbial molecules. This high reactivity implies mechanisms preventing inappropriate activation of pro-inflammatory immune signals that can otherwise favours cancer progression or induce tissue damages such as those occurring in auto-immune diseases. We used Drosophila flies model system for the identification of new enzymes of the ubiquitin specific protease family regulating evolutionary conserved immune signals in both Drosophila and Humans. These enzymes specifically control the stability or the activated status of target proteins by hydrolysing a small peptide, called ubiquitin, which is linked as a monomer or polymers on protein. Moreover, they constitute promising and yet poorly explored targets for the finding of drugs useable as therapeutic molecules. We found a set of three ubiquitin specific proteases regulating immune signals in response to infections, among which USP2 controls the homeostasis of an essential signalling component -named Imd- by promoting its degradation at the proteasome. Beyond the immune response, this work highlights how a specific enzyme, USP2, may regulate its targets in physio-pathological processes for a better understanding of its pathogenic activity in cancers and inflammation.
Background
Conjugation of ubiquitin monomers or polymers to proteins is a key mechanism for controlling their activity or stability [[1]]. Lysine (Lys) residues of proteins can be modified by a single ubiquitin monomer or by polymers of ubiquitin (polyubiquitin) each linked through Lys 48 (K48) or through Lys63 (K63) of the ubiquitin molecule, or by other kinds of polymers. Whereas K48-linked polyubiquitin (UbK48) mainly triggers degradation of proteins by the proteasome, monoubiquitination, K63-linked polyubiquitin (UbK63) and other polyubiquitin chains regulate the activity, the conformation or the subcellular localisation of proteins [[2],[3]]. Mammalian genomes contain about one hundred ubiquitin proteases – the enzymes that remove ubiquitin moieties from proteins- that are divided in five subfamilies, among which the Ubiquitin Specific Proteases (USPs) subfamily represents the major class in both human and Drosophila [[4]-[7]]. The NF-κB dependent signalling pathways, that are central to pro-inflammatory and immune signalling, are tightly regulated by the ubiquitination of several of their protein components including RIP1, interacting with the tumour necrosis factor receptor 1 (TNF-R1), and downstream protein kinase complexes [[5],[8]]. A growing number of ubiquitin proteases have been found to mediate transient inhibition of NF-κB- pathways frequently sharing a same target which raises the question of their specificity and functional interrelationships [[5]].
In Drosophila, two conserved NF-κB-like signalling pathways, Toll and Imd, contribute to innate immunity by promoting the expression of antimicrobial peptide (AMP) encoding genes -mainly in fat body cells- in response to infections [[9]]. The Toll receptor is activated by binding of the processed cytokine Spätzle (Spz), whose cleavage depends on upstream extracellular proteolytic cascades initiated by circulating peptidoglycan recognition proteins (PGRPs) following the recognition of various pathogens associated patterns, such as the Lys-type peptidoglycans from Gram-positive bacteria [[10],[11]]. Activated Toll associates with adaptor proteins dMyd88 and Tube, ultimately resulting in the activation of the NF-κB like factors, Dif and/or Dorsal (DL), which activate the transcription of a set of AMP encoding genes including Drosomycin (Drs), immune induced molecule 1 (IM1) and Attacin (AttA) [[10],[12]-[14]]. The Imd pathway is induced by the direct binding of diaminopimelic acids containing bacterial peptidoglycan fragments to the transmembrane receptor PGRP-LC/Ird7 and results in the activation of another set of AMPs encoding genes, including Diptericin (Dpt), Defensine (Def) and Attacin A (AttA) [[12]]. PGRP-LC associates with the cytoplasmic scaffolding protein Imd [[15]] which mediates the activation of Tak1 and Kenny (Key), the Drosophila IKKγ homolog, resulting in the phosphorylation of the NF-κB like factor Relish (Rel) [[16]].
We have previously demonstrated that Imd is ubiquitinated by UbK63 and that the ubiquitin specific protease USP36/Scny negatively regulates signal transduction by hydrolysing UbK63 from Imd [[17]]. Complex regulation of immune signals by ubiquitin-dependent mechanisms prompted us to identify other ubiquitin specific proteases (USPs) acting in Imd and Toll dependent immune signalling. To this end, we have designed and screened an RNAi library targeting the Drosophila USPs in S2 cells. We report here the identification of three regulators of the Imd pathway: the already described USP36 and two new USPs, USP2 and USP34/Puf (Puffyeye [[18]]), and of one regulator of the Toll pathway: USP34. We demonstrate that USP2 and USP34 are required to prevent constitutive activation of immune signalling in vivo. However, while USP2 is also required to prevent excessive activation of the Imd pathway in infected flies, USP34 displays differential requirement depending on the antimicrobial peptide gene analysed: silencing Usp34 enhanced the activation of AttA, Drs and IM1. In the opposite, USP34 is required for full activation of the two specific Imd-dependent genes Dpt and Def in response to Gram-negative bacteria. These results suggest a complex requirement of USP34 in these two pathways. Focusing on USP2 biochemical function, we show that USP2 binds to Imd and promotes the cleavage of UbK48 chains from the protein in both cultured cells and flies. Surprisingly, USP2 also targets Imd for degradation, a function which, from our proteomic analysis of USP2 partners and subsequent co-immunoprecipitation experiments, likely occurs at the level of the proteasome. Interestingly, UbK48 chains linkage on Imd, acting as a tag for its proteasomal degradation, is fully prevented in response to infection therefore ensuring the required protein stabilisation for signal transduction. Thus, Imd homeostasis is tightly regulated by ubiquitination and USP2 controls Imd pathway activation by regulating the steady-state level of the Imd protein.
Results
Three USPs regulate the Imd pathway in S2 cells
We created a double strand RNA interference (dsRNA) library targeting USPs encoded in the Drosophila genome (Additional file 1: Table S1) (see also [[7],[19]]). This library was screened in Drosophila S2 cells on PGRP-LC-dependent activation of Imd pathway mediated by adding heat-killed Escherichia coli (E. coli) in the culture medium. Activation of the pathway was monitored by using an Attacin-luciferase (AttA-luc) reporter gene and the signal was normalised to that from a control Actin-luciferase (Act-luc) reporter gene [[20]]. Silencing Usp2, Usp34 and Usp36 genes resulted in over-activation of the Imd dependent AttA promoter whereas silencing the other Usp genes had no significant effect (Figure 1A). Constitutive activation of the AttA promoter was also detected when the same three Usp genes were silenced in the absence of heat-killed E. coli (Figure 1B). Monitoring Usp2, Usp34 and Usp36 transcripts in RNAi-treated cells showed effective gene silencing (Additional file 2: Figure S1). Activation of the Imd pathway in Usp2, Usp34 and Usp36 silenced cells was strictly dependent on the NF-κB like protein Rel since co-silencing Rel with each of these three genes fully prevented activation of the AttA promoter (Figure 1C). Co-silencing Imd with either Usp2 or Usp34 strongly decreased the induction observed in only Usp2- or Usp34-silenced cells indicating a crucial contribution of Imd (Figure 1C). Finally, as expected from our previous findings demonstrating that USP36 directly targets Imd [[17]], we observed no induction of AttA when Imd and Usp36 were co-silenced in S2 cells (Figure 1C). This screen thus defines USP2, USP34 and USP36 as inhibitors of the Imd pathway in S2 cells.
Figure 1.
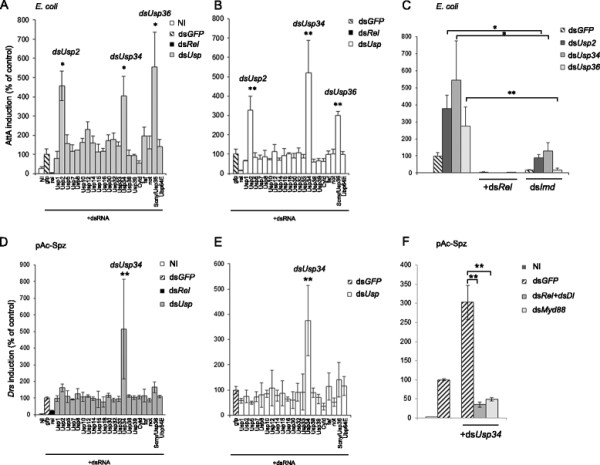
Screening USPs regulating Imd and Toll pathways.Drosophila S2 cells were co-transfected with the indicated dsRNAs, pAct-luc normaliser and either the pAttA-luc(A-C) or the pDrs-luc(D-F) reporter gene. A,C: The Imd pathway was activated by adding heat-killed E. coli. D,F: The Toll pathway is stimulated by co-transfecting cells with pAc-Spz. B,E: Cells are not stimulated (ie, neither by E. coli(B) not by pAc-Spz (E)) in order to look after constitutive deregulation of the pathway. A,D,C,F. NI: not induced control. A-F. Histograms represent the % of induction compared to double strand gfp treated control cells. One out of three independent experiments is shown. Error bars indicate standard deviation of technical triplicates. Significant differences (Student-t-test) p < 0.05 (*), p < 0.02 (**).
USP34 inhibits the Toll pathway in S2 cells
To identify ubiquitin proteases regulating the Toll pathway, we activated this pathway by expressing Spz -the ligand of Toll- in S2 cells. Expression of a Drs-luc reporter construct was monitored and normalized to that of an Act-luc construct [[21]]. A preliminary assay indicated that the NF-κB factors Dorsal (Dl) and Rel, but not Dif, were required for the Toll-mediated activation of the Drs promoter in S2 cells (Additional file 2: Figure S2) as also described in a previous study [[22]]. Silencing upstream components of the pathway, dMyd88 or Spz, prevented the activation of the Drs promoter as expected (Additional file 2: Figure S2). By contrast, silencing upstream components of the Imd pathway, PGRP-LC, Imd, Tab2 and Key, did not affect expression from the Drs promoter induced by Spz, indicating that Rel is the sole Imd pathway component contributing to Toll activation in S2 cells (Additional file 2: Figure S2). Screening the RNAi library in Spz-expressing cells revealed that Usp34 silencing resulted in a significant enhancement of Drs promoter activity when compared to control cells (Figure 1D). Similarly, Usp34 was the only gene whose silencing induced the Drs promoter in cells not expressing Spz (Figure 1E). This activation was strictly dependent on downstream NF-κB factors (Rel and Dl) and on the adaptor protein dMyd88 (Figure 1F). We therefore identified USP34 as a new inhibitor of the Toll pathway both in stimulated and un-stimulated cells.
Overexpressing USP2 or USP34 suppresses fly immunity in vivo
The USP36 negative function on flies resistance to infection and antimicrobial peptide expression has been previously described [[17]]. To assess the function of USP2 and USP34 on immune signalling in vivo, we created a UAS-Usp2 transgenic strain and used a P[UAS] insertion located upstream of the Usp34 transcription unit, P[EPgy2]ash2EY03971 (hereafter referred to as UAS-Usp34), in order to overexpress each corresponding genes under the control of the GAL4 transcription factor [[23]]. Overexpression of these two genes was induced in adults through the heat-shock HspGal4 driver line and was verified by quantitative Real Time PCR at 3, 6, 9 and 12 hours post-heat shock (Additional file 2: Figure S3). A strong diminution of Dpt expression was observed in E. coli infected flies overexpressing USP34 and to a lesser but significant extent, in flies overexpressing USP2 (Figure 2A). Inhibition of AMP gene expression was associated with increased fly sensitivity to the Gram-negative pathogen Enterobacter cloacae (Figure 2B) and Klebsellia pneumonia (not shown). Similarly as observed for Dpt, overexpressing USP34 significantly reduced Drs expression at 12 and 24 hours after infection with the non-pathogenic Gram-positive bacteria Micrococcus luteus (Figure 2C). This phenotype was associated with increased fly sensitivity to the Gram-positive pathogen Enterococcus faecalis (Figure 2D). Therefore, when overexpressed in vivo, USP2 and USP34 behave as negative regulators of the fly immune response to bacterial infections.
Figure 2.
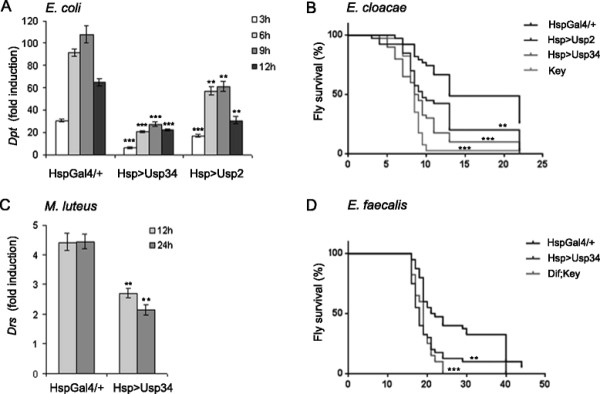
Overexpressing USP2 or USP34 suppresses fly immunityin vivo.A-D. Flies were heat shocked 12 hours before infection. A. Quantitative analysis of Dpt mRNAs levels by RT-qPCR at 3, 6, 9 and 12 hours post infection with E. coli. B. Forty heat-shocked flies of 5 days old were infected with E. cloacae and their survival kinetics was followed over 24 hours. The mutant key is used as a Gram-negative bacteria sensitive control (non-heat shocked flies). C. Quantitative analysis of Drs mRNAs by RT-qPCR at 12 and 24 hours post infection with M. luteus. D. Forty flies of 5 days old were infected with E. faecalis 6 hours following heat shock treatment and their survival kinetics was followed over 48 hours. The mutant w1118;Dif,Key is used as a Gram-positive bacteria sensitive control (non-heat shocked flies). A,C: Results are expressed as the fold induction level compared to the level of Dpt(A) or Drs(C) mRNAs in non-infected flies. Error bars indicate standard deviation between technical replicates. One representative experiment out of three is shown. Significant difference: p < 0.05 (*), p < 0.002 (**) or p < 0.0001 (***) compared to HspGal4/+control flies (Student’s-t-test). B,D: Results are expressed as % of surviving flies. One representative experiment out of three is shown. Significant difference: p < 0.002 (**) or p < 0.0001 (***) compared to HspGal4/+control flies (Log-rank (Mantel-Cox) test). A-D. Flies’ genotypes are: w1118;HspGal4/+ (HspGal4/+), w1118;HspGal4/+; P{EPgy2}ash2EY03971/+ (Hsp > Usp34), w1118;HspGal4/+;UAS-Usp2/+ (Hsp > Usp2), w1118;key1 (key) and w1118; Dif1, key1(Dif,key).
USP2 and USP34 prevent constitutive immune signalling in vivo but differentially control antimicrobial peptide genes expression following bacterial infection
To investigate further the functions of USP2 and USP34 in vivo, we created transgenic fly lines expressing inverted repeats (IR) of Usp2 (Usp2-IR #5M hereafter designed as Usp2-IR) or Usp34 (Usp34-IR #1M hereafter designed as Usp34-IR) inducing efficient gene silencing in the living flies (Additional file 2: Figure S4A,B). Specific silencing of either Usp2 or Usp34 in the adult fat body was then achieved by using the driver line c564 [[24]] and resulted in a significant constitutive activation of Dpt and AttA in the absence of immune challenge (Figure 3A). We obtained similar results with other transgenes carrying different silencing sequences designed to silence the same two genes from the VDRC collection [[25]] (Additional file 2: Figure S4A-D). In addition, a significant three-fold activation of two Toll-pathway target genes Drs and IM1 was also observed in Usp34-silenced flies (Figure 3A). Silencing Imd strongly reduced the constitutive activation of Imd-dependent target genes AttA and Dpt induced by silencing either Usp2 or Usp34 indicating that Imd contribute to the observed up-regulation of these antimicrobial genes (Figure 3B). Moreover, enhanced activation of Dpt and AttA expression was observed in Usp2-silenced flies three hours after a septic injury with E. coli and this activation was fully prevented in Imd-silenced flies (Figure 3C). Enhanced activation of Dpt and AttA in Usp2-silenced flies was similarly observed at several time-points from 3 to 9 hours following infection (Additional file 2: Figure S5A).
Figure 3.
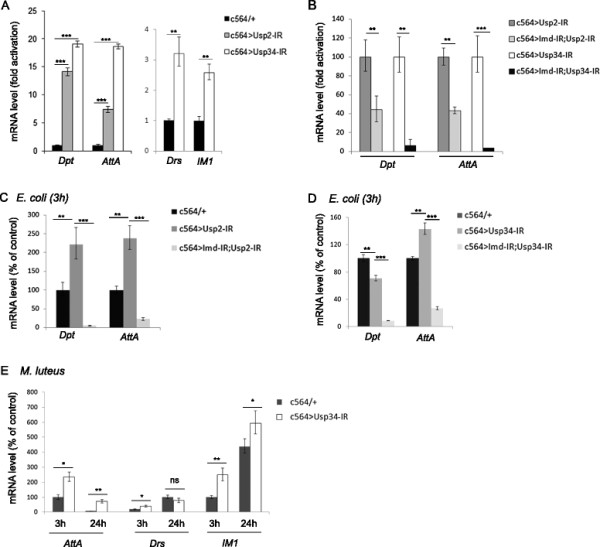
USP2 and USP34 regulate immune signallingin vivo. Quantitative analysis of Dpt, AttA, Drs and IM1 mRNAs by RT-qPCR. A. Histograms present the fold activation of the indicated gene mRNA in Usp2 (c564 > Usp2-IR) or Usp34 (c564 > Usp34-IR) silenced flies compared to control flies (c564Gal4/+) raised in similar conditions. B. Dpt and AttA induction in double silenced flies (c564 > Usp2-IR, Imd-IR or c564 > Usp34-IR, Imd-IR) expressed as a % of activity of Usp-only silenced flies (c564 > Usp2-IR or c564 > Usp34-IR, respectively). C,D. Activation of Dpt and AttA in Usp2- (C) or Usp34(D) -silenced flies, in combination or not with an Imd-silencing transgene-, at 3 hours post E. coli infection, expressed as a % of activity compared to infected control flies (c564/+). E. Activation of AttA, Drs and IM1 in Usp34-silenced flies at 3 and 24 hours post M. luteus infection expressed as a % of activity compared to infected control flies (c564/+) (100% is the maximal activation observed in control flies at 3 hours post infection for AttA, and IM1 and at 24 hours post-infection for Drs). A-E. One representative experiment out of three is shown. Error bars indicate standard deviation between technical triplicates. Significant differences (t-test), p < 0.05 (*), p < 0.02 (**), p < 0.001 (***).
In contrast, antimicrobial peptide gene induction upon bacterial infection was differentially modified in Usp34-silenced flies. Indeed, in flies infected by E. coli, the induction of AttA was enhanced in Usp34 silenced flies compared to control flies (Figure 3D, Additional file 2: Figure S5C). However, the induction of Dpt expression was significantly compromised in Usp34-silenced flies, resulting in a 30 to 50% reduction of Dpt induction from 3 to 9 hours following infection by E. coli (Figure 3D, Additional file 2: Figure S5C). The expression of Defensin C (Def) an additional Imd-dependent antimicrobial peptide encoding gene, was also significantly deregulated in both Usp34-silenced non-infected flies and at 3 hours post-infection (Additional file 2: Figure S5B,C) but then, it was strongly compromised from 6 to 9 hours following E. coli infection (Additional file 2: Figure S5C). Finally, when the Toll pathway was specifically activated by infecting flies with M. luteus, all three target genes analyzed, AttA, Drs and IM1 were significantly enhanced in Usp34 silenced flies compared to control flies at 3 hours following the infection, while at 24 hours infection, up-regulation was significant only in the cases of AttA and IM1 (Figure 3E).
These data suggest that in non-infected flies, both USP2 and USP34 are required in fat body cells to inhibit immune signals. However, in infected flies, USP2 is acting as a negative regulator but USP34 is differentially required for either the activation or the inhibition of antimicrobial peptide genes.
Finally, the monitoring of Usp2 and Usp34 mRNAs in response to E. coli or M. luteus infections revealed no major change in Usp2 or Usp34 gene expression (below 1,6 fold change) (Additional file 2: Figure S6). This suggests that the activity or stability of these two proteases in response to infection is mainly regulated at the protein level.
USP2 interacts with Imd and promotes cleavage of Imd-linked UbK48
We decided to focus on the function of USP2 in the Imd pathway. So, we transfected USP2-Myc and Imd-V5 tagged expressing constructs in S2 cells to assess whether the corresponding proteins interact with each other (Figure 4A). Indeed, we observed co-immunoprecipitation of the two proteins indicating that USP2 and Imd belong to a same protein complex (Figure 4B). Imd is a scaffolding protein containing a death domain located in the C-terminus part of the protein while its N-terminus part is required for its interaction with PGRP-LC [[15],[26]]. Using V5 tagged Imd truncated constructs (Figure 4A), we showed that USP2 preferentially interacts with the PGRP-LC-interacting Imd-N-ter part in S2 cells (Figure 4B). GST pull-down assays using GST-tagged full-length and truncated forms of Imd expressed as recombinant proteins in bacteria confirmed this preferential interaction (Additional file 2: Figure S7).
Figure 4.
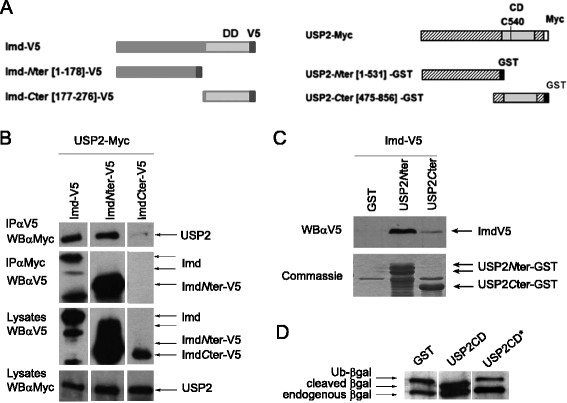
USP2 interacts with Imd. A. Representation of the constructs used in this study. DD: death domain in Imd, CD: catalytic domain in USP2, V5: V5 tag, Myc: Myc tag. Numbers in brackets indicate amino-acid position. B. Drosophila S2 cells were cotransfected with the expression constructs encoding USP2-Myc and either Imd-V5, or Imd-N-ter-V5 or Imd-C-ter-V5 as indicated. Cells lysates were coimmunoprecipitated (IP) with either anti-V5 or anti-Myc antibodies and analysed by western blot (WB) with either anti-Myc or anti-V5 antibodies as indicated. C. S2 cells were transfected with the expression construct encoding Imd-V5 and lysed after 48 h. Cells lysates were pre-cleared and subjected to GST pull down assays using GST fusion proteins with either the USP2-N-ter [1–531] or the USP2-C-ter [475–856] - or with GST alone (GST). Gel was coloured with Coomassie to visualize GST-fusion proteins in the input (bottom part). D. GST-fusion of the wild type (USP2CD) or mutated (USP2CDC540S indicated USP2CD*) catalytic domain of USP2 were coexpressed with Ub-β-gal for 4 hours at 28°C in transformed E. coli XL1 Blue. Substrate cleavage was analysed by western blotting with anti-βgal antibodies.
The catalytic domain of USP2 - containing the critical cysteine residue required for ubiquitin chain hydrolysis at position 540- is located in the C-terminal part of the enzyme [[27]] (Figure 4A). By using GST pull-down assays with GST-tagged full length or truncated forms of USP2 (USP2-N-ter [AA:1–531] and USP2-C-ter [AA:475–856]), we finally showed that USP2 preferentially interacts with Imd through its non-catalytic N-ter domain (Figure 4C).
We then expressed the USP2 -C-ter catalytic domain (USP2CD) in bacteria expressing a fusion protein of ubiquitin with β-galactosidase (Ub-β-gal), a substrate for deubiquitination. We observed a clear cleavage of this substrate indicating that Drosophila USP2 is an active deubiquitinating enzyme (Figure 4D). Mutation of the conserved Cys540 to Ser (C540S) resulted in no hydrolysis of the Ub-β-gal fusion protein (Figure 4D, USP2CD*).
To test the ability of USP2 to deubiquitinate Imd, we co-expressed full-length USP2 with Imd in S2 cells. Whereas the level of UbK63-Imd was not modified by the presence of wild type or catalytically inactive full length USP2, the amount of UbK48-Imd was reduced in cells overexpressing USP2 but not in cells expressing its catalytically inactive form in both untreated or MG132 treated cells (Figure 5A). This suggests that USP2 specifically hydrolyses Imd-linked UbK48. Consistent with this hypothesis, UbK48-Imd accumulated in Usp2-silenced cells both in MG132 treated or untreated cells (Figure 5B).
Figure 5.
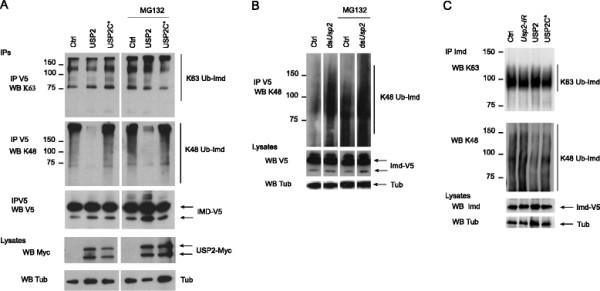
USP2 hydrolyses Imd-linked UbK48. A. Drosophila S2 cells were cotransfected with Imd-V5 and either wild type USP2 or mutant USP2C540S (USP2C*) expressing constructs in untreated cells or cells treated with MG132 at 20 μM for 4 hours before cell lysis. Imd-V5 was immunoprecipitated with anti-V5 antibodies (IP V5) and Imd-ubiquitinated forms were detected by western blot with either anti-UbK63 (WB K63) or anti-UbK48 (WB K48) antibodies as indicated. Imd full length was detected with anti-V5 antibodies (IP V5 WB V5). Expression of USP2 was assessed by western blot of whole cell lysate with anti-Myc antibodies (WB Myc). B. Drosophila S2 cells were cotransfected with Imd-V5 in the presence or not of Usp2 silencing dsRNA (dsUsp2). Imd-V5 was immunoprecipitated with anti-V5 antibodies and Imd-ubiquitinated forms were detected by western blot with anti-UbK48 antibodies (IP V5 WB K48). Imd full length was detected in the cell lysate with anti-V5 antibodies (WB V5). C. Endogenous Imd was immunoprecipitated with anti-Imd antibodies from extracts of c564/+ control flies (Ctrl), or flies expressing either the Usp2 silencing transgene (Usp2-IR), or the wild type (USP2) or the catalytically inactive form (USP2C*) of USP2, under the control of the c564Gal4 driver line (IP Imd). Ubiquitinated forms of Imd were detected with either UbK63 (WB K63) or UbK48 (WB K48) antibodies as indicated. The amount of endogenous Imd proteins was detected in total fly extracts with Imd antibodies (WB Imd). A-C. Anti-tubulin antibodies served as loading control (WB Tub).
To characterize the activity of USP2 on Imd-linked ubiquitin chains in vivo, we immunoprecipitated the endogenous Imd protein from flies extracts. Silencing Usp2 resulted in the accumulation of UbK48-Imd (Figure 5C). On the opposite, expressing USP2, but not the catalytic mutant, resulted in a strong diminution of immunoprecipitated UbK48-Imd compared to control flies (Figure 5C). As observed in S2 cells, UbK63-Imd chains were not affected by Usp2 gene extinction or overexpression (Figure 5C). Therefore, we conclude that Drosophila USP2 is an active deubiquitinating enzyme specifically promoting the hydrolysis of UbK48 linked to Imd.
USP2 promotes Imd degradation by the proteasome
Our data indicate that Imd is permanently linked by UbK48 putatively ensuring its turnover through proteasomal degradation. Indeed, inhibiting the proteasome in S2 cells with MG132 led to the accumulation of full-length and cleaved Imd when compared to untreated control cells (Figure 6A). Remarkably, silencing Usp2 also provoked a clear accumulation of full length and cleaved Imd compared to control cells reaching similar amount as in MG132 treated cells (Figure 6A). Likewise, silencing of Usp2 in the fat body resulted in the accumulation of endogenous Imd (Figure 6B). This is surprising because we have previously shown that USP2 promotes the hydrolysis of UbK48 linked to Imd, a process which is expected to save Imd from proteasomal degradation. On the opposite, our results clearly indicate that Usp2 silencing induces a blockade in the Imd degradation process thus suggesting that USP2 is required for Imd degradation. By contrast, the amount of the Toll pathway component Cactus was not modified by silencing Usp2 in S2 cells (Figure 6C).
Figure 6.
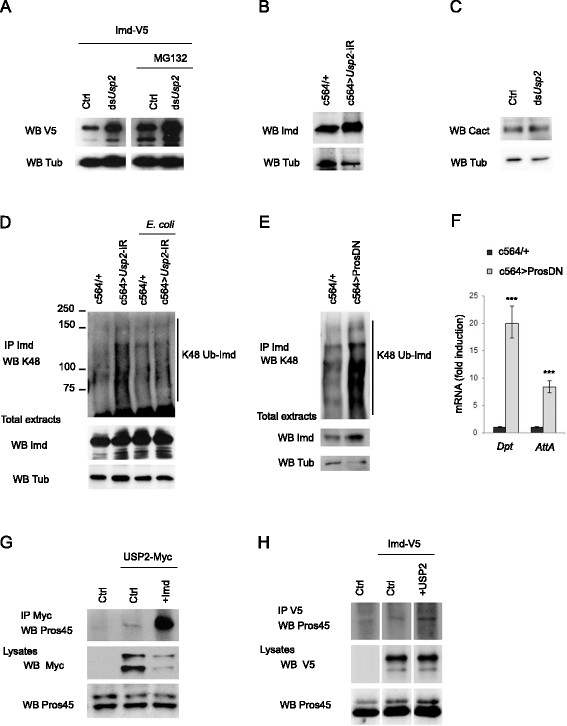
Imd proteasomal degradation requires USP2. A. S2 cells were transfected by Imd-V5 expressing construct and treated or not by Usp2 silencing dsRNAs (dsUsp2). Cells were incubated or not with MG132 at 20 μM. Cell lysates were immunoblotted with anti-V5 antibodies (WB V5). B. Protein extracts of c564/+ control or c564/Usp2-IR flies were immunoblotted with anti-Imd antibodies (WB Imd). C. S2 cells lysates treated or not with dsUsp2 were immunoblotted with anti-Cact antibodies (DSHB) (WB Cact). D. Endogenous Imd was immunoprecipitated from extracts of c564/+ or c564/Usp2-IR flies infected or not by E. coli. Ubiquitinated forms of Imd were detected with anti-UbK48 antibodies (IP Imd WB K48). Total extracts were immunoblotted with anti-Imd antibodies (WB Imd). E. Endogenous Imd was immunoprecipitated from extracts of c564/+ or c564/Pros26[[1]]; Prosbeta[[1]]/+ flies (c564 > ProsDN). Ubiquitinated forms of Imd were detected by anti-UbK48 antibodies (IP Imd WB K48). A-E: Anti-tubulin immunoblots served as loading control (WB Tub). F. Quantitative analysis of Dpt and AttA mRNAs from c564 > ProsDN compared to c564/+ control flies. Histograms present the fold activation of each mRNA. Error bars indicate standard deviation between technical triplicates. Significant differences with control (t-test) p < 0.001 (***). G. S2 cells were cotransfected with pAc-USP2-Myc and either pAc-Imd or a control empty plasmid (Ctrl). USP2 was immunoprecipitated with anti-Myc antibodies and immunoblotted with anti Pros45 antibody (DHSB) (IP Myc WB Pros45). Cells lysates were immunoblotted with anti-Myc and anti Pros45 antibodies (WB Myc, WB Pros45). H. S2 cells were cotransfected with pAc-Imd-V5 and either pAc-Usp2 or a control empty plasmid (Ctrl). Imd was immunoprecipitated with anti-V5 antibodies and endogenous Pros45 was detected (IP V5 WB Pros45). Whole cells lysates were revealed with anti-V5 and anti Pros45 antibodies (WB V5 WB Pros45).
Since a proper immune response would require stable Imd, we investigated whether bacterial infection modifies the amount of UbK48-linked and full length Imd. Strikingly, UbK48-Imd accumulation seen in Usp2-silenced flies was strongly reduced in infected flies compared to uninfected flies indicating that the linkage of UbK48 on Imd is actively prevented in response to immune challenge, thus allowing for Imd stabilisation (Figure 6D). To investigate whether accumulation of Imd protein could be sufficient to induce ectopic activation of the Imd pathway in vivo, we blocked proteasome function in flies expressing two conditional dominant-negative proteasome subunits (UAS-pros261ts; UAS-prosβ21ts) [[28]]. Flies kept at the restrictive temperature (30°C) for 72 hours accumulated UbK48 –linked and full length Imd (Figure 6E) and concomitantly, they displayed significant activation of the AttA and Dpt expression (Figure 6F). Altogether, our results suggest an essential role of USP2 in both the hydrolysis of UbK48 linked to Imd and the proteasomal degradation of Imd thus preventing its accumulation and subsequent constitutive activation of the Imd pathway.
USP2 and Imd synergistically interact with the proteasome
To investigate how USP2 might ensure its regulatory function on Imd homeostasis, we realised a pull-down assay using S2 cell lysate and GST or GST-USP2 as baits to identify USP2-interacting proteins. Eluates were then analysed by a tandem mass spectrometry-based proteomic approach. Among other candidates specifically identified as associated with GST-USP2 (Additional file 3: Table S2), the proteasomal subunit 8 (Pros45) may trigger USP2 binding to the proteasomal machinery. Indeed, endogenous Pros45 co-immunoprecipitate with USP2-Myc (Figure 6G). Interestingly, a much higher amount of this proteasomal subunit was detected in the presence of overexpressed Imd (Figure 6G). Reciprocally, while Pros45 was hardly detectable in the purified product from immunoprecipitated Imd-V5, it was clearly observed when USP2 was overexpressed (Figure 6H). Thus, the Imd–USP2 complex apparently binds more efficiently to the proteasome than do Imd or USP2 alone. Taken together, our data suggest that USP2 and Imd synergistically bind to the proteasome and that USP2 contributes to Imd degradation at the proteasome level.
Discussion
By using an RNAi screening approach, we demonstrate here the importance of three ubiquitin proteases, USP2, USP34 and USP36, in the regulation of the Imd immune signalling pathway. In addition, USP34 was identified as the only negative regulator of the Toll pathway in this screen. While this study and previous work [[17]] show a clear negative regulatory function for USP2 and USP36 on the Imd pathway in both un-infected and infected flies, USP34 can have both negative, (i.e., on AttA, Drs and IM1) and a positive (i.e., on Dpt and Def), regulatory functions on antimicrobial peptide genes in infected flies. Similar differential effect on various antimicrobial peptide genes induction has been observed with the POSH E3 ligase targeting TAK1 [[29]] while only a subset of Imd target genes (i.e., Dpt and Dro but not AttA and CecA1) are affected in a faf (fat facets) mutant, where faf encodes a deubiquitinating enzyme targeting Imd [[30]]. Our result suggest a complex requirement of USP34 putatively acting at different levels of these two pathways. In unchallenged conditions, USP34 may interfere with common negative regulatory mechanisms preventing ectopic transcriptional activation of NF-κB target genes while in challenged conditions, USP34 may target components that are differentially required in the two pathways.
Focusing on the function of USP2, we found that it interacts with Imd and specifically promotes the hydrolysis of UbK48 from the protein, thus displaying different substrate specificity than USP36 which was previously described to promote the hydrolysis of UbK63 from Imd [[17]]. Whereas the linkage of UbK63 on Imd activates signal transduction [[17],[31]], we show here that UbK48 serves as a tag for Imd degradation by the proteasome. Thus, differential ubiquitination of Imd regulates either its activity or its stability and is tightly controlled by the complementary activity of USP36 and USP2 (Figure 7). Even though they target different ubiquitin chains and have different effects on Imd, both enzymes contribute to down-regulation of the Imd pathway.
Figure 7.
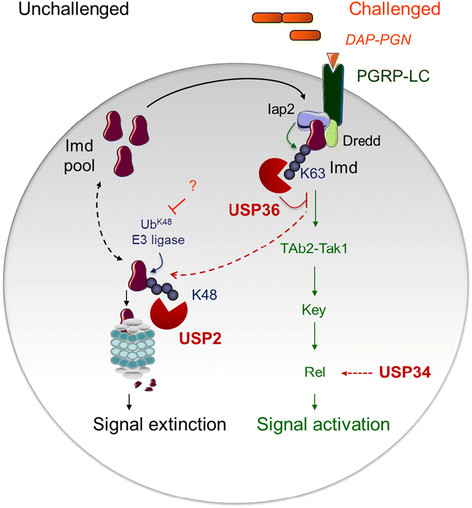
(Summary illustration): Three USPs regulate Imd activation or stability and downstream signalling. Three ubiquitin specific proteases regulate the Imd pathway, USP34 is putatively acting at the level of NF-κB like factors associated complexes (Rel in the case of Imd pathway) while USP2 and USP36 differentially targets the scaffolding molecule Imd by hydrolysing UbK48 (this study) and UbK63 [[17]] chains, respectively. Imd is subjected to a permanent turn-over via the linkage of UbK48 ubiquitin chains and subsequent proteasomal degradation. This prevents Imd accumulation and constitutive signal activation in unchallenged conditions. The deubiquitinating enzyme USP2 is required for both UbK48 hydrolysis and Imd degradation at the level of the proteasome. In response to an immune challenge, the activity of the E3 ligase would be prevented resulting in the observed Imd stabilisation and accumulation. It has been previously described that Imd activation in response to immune challenge results in its cleavage by Dredd and linkage of UbK63 by Iap2 [[31]]. USP36 deubiquitinates Imd linked UbK63 thus preventing inappropriate or excessive signal transduction and putatively promoting ubiquitin chain editing through the replacement of activating UbK63 by UbK48 degradative chains [[17]].
Actually, it was first rather surprising that while USP2 promotes the cleavage of UbK48 from Imd, its physiological function is not to rescue Imd from degradation but rather to promote its degradation by the proteasome. This was clearly demonstrated by using silencing experiments: in Usp2-silenced cells, or in flies expressing the Usp2-IR silencing transgene, UbK48 and full-length Imd accumulated indicating a blockade of the Imd degradation process. At the molecular level, the simplest explanation for this result is that USP2 drives the UbK48-Imd molecules to the proteasome and/or ensures the removal of UbK48 from Imd at the level of the proteasome, then allowing for Imd degradation by the proteasome machinery (Figure 7). Indeed, the release of attached polyubiquitin chain is necessary to permit the entry of a specific substrate to the proteasomal cavity [[32],[33]]. Moreover, it has been recently illustrated that deubiquitination by the yeast enzyme Ubp3 –which interacts with the 26S proteasome subunit- can either save proteins from destruction or facilitate protein destruction by the 26S proteasome, depending on stress conditions and Ubp3 amount in the cell [[34]]. Our evidence that USP2 interacts with Pros45, the 26S subunit 8 of the proteasome, and that this interaction is significantly enhanced in the presence of Imd supports similar essential role of USP2 in Imd degradation at the level of the proteasome. Interestingly, artificial overproduction of USP2 by tissue-directed or heat-shocked-induced expression of the Usp2 expressing transgene in non-infected flies, also leads to Imd accumulation (data not shown). Therefore, as shown for Ubp3, depending on its total amount, USP2 may either save a target protein from degradation by the proteasome or favour its degradation.
The accumulation of Imd in Usp2-silenced flies most probably contributes to the observed constitutive induction of Imd-dependent AMP gene expression. Consistent with this, blocking the proteasome by expressing dominant-negative proteasome subunits in transgenic flies similarly resulted in accumulation of full length Imd concomitantly with constitutive activation of the Imd pathway. The molecular mechanisms underlying ectopic activation of the Imd pathway, when Imd is overexpressed or inefficiently degraded, are not known. Notably, it should be assess in the future whether it is associated or not with increased Imd K63-ubiquitination. Actually, in the absence of immune challenge that would activate the E3 ligase Iap2, which is responsible for the linkage of K63 chains on Imd [[31]], Imd accumulation and self-aggregation may favour the self-assembly of activating complexes independently of the linkage of ubiquitin moieties. Interestingly, similar activation of Dpt expression was previously observed following proteasomal inhibition and was associated with Rel stabilisation [[35]]. Thus, proteasome activity, by acting on several components of the pathway (at least Imd and Rel), likely has an important function in damping down immune signalling in the absence of infection.
Remarkably, the accumulation of UbK48 -Imd seen in Usp2-silenced flies mostly disappeared following an immune challenge. Subsequently, Imd is significantly stabilised in challenged flies compared to unchallenged flies. Since Usp2 was silenced in these flies, this disappearance of UbK48 -Imd cannot be a consequence of USP2 deubiquitinating activity. Rather, we suggest that infection inhibits the linkage of UbK48 to Imd by an unidentified E3 ubiquitin ligase and/or activate another DUB that would cleave K48 ubiquitin chains linked to Imd.
The human homologs of USP2 and USP34 also play regulatory roles in NF-κB-dependent immune signalling. One study reported that USP34 is a regulator of T-cell receptor (TCR)-dependent lymphocyte activation where it acts as a down-regulator of NF-κB signalling [[36]]. Although its molecular target remains to be identified, USP34 functions downstream of the IKK complex in human cells, putatively on proteins that would be commonly required in these different NF-κB signalling pathways. The fact that USP34 is required in the two NF-κB immune signalling pathways Toll and Imd also argues in favour of USP34 targeting common regulatory mechanisms at the level of NF-κB transcription factors. This hypothesis is reinforced by the observation that in response to infection, USP34 differentially regulates AMP genes expression which may typically reflect differential activity at the level of NF-κB-containing complexes. Of note, however, co-silencing of the scaffolding proteins Imd or dMyd88 fully disrupted the PGRP-LC- or the Toll-dependent activation of immune signals observed in Usp34-silenced condition, respectively. This suggests that these upstream molecules are required to ensure a minimal activation of NF-κB-like factors that are then tightly regulated by USP34-dependent mechanisms that stay to be discovered in both Drosophila and mammalian models.
A set of different studies point to a regulatory function of the mammalian USP2a isoform on TNF-R1 dependent activation of the NF-κB pathway. First, silencing Usp2a contributes to TNFα-dependent hepatocyte survival in mice, presumably due to enhanced transcriptional activity of NF-κB [[37]]. Second, USP2a hydrolyses UbK63 from RIP1 thus preventing NF-κB activation while promoting caspase-mediated cell death in response to TNFα stimulation in human cells [[38]]. Two other studies, by contrast, provide controversial results by describing a positive role for USP2a in TNF-R1 dependent NF-κB signalling [[39]] or in the regulation of TCR-induced NF-κB activation [[40]]. These discrepancies may be due to dose-dependent effects of USP2 on multiple targets.
Conclusion
In addition to the NF-κB pathway, a huge and increasing number of proteins and pathways seems to be controlled by USP2, which is also deregulated in many cancers [[41]-[46]]. Our demonstration of USP2 involvement in the proteasomal degradation of Imd suggests a more extended function at the proteasome level than previously anticipated. It may highlight new mechanisms of action of USP2 on its targets and provide some molecular explanations for its tumorigenic function in humans in the future.
Methods
Cell culture and RNA silencing
Drosophila S2 cells were maintained in Schneider’s Drosophila medium supplemented with 10% heat-inactivated serum (FCS, Invitrogen). Gene inactivation was achieved by incubating 0.4 μg double strand RNA (dsRNA) for 48 h at 26°C with 1.2×105 S2 cells cultured in 96-well microplates (adapted from [[47]]. DNA templates for dsRNA synthesis were generated by PCR (MEGAscript RNAi kit, Ambion) using the primers designed from Heidelberg Fly Array RNAi libraries (www.genomernai.org/). To monitor Imd and Toll pathways activation, we used the reporter constructs pAttA-luc [[48]] and pDrs-luc [[21]], respectively. We further constructed a normaliser pAct-luc (Promega). DNA transfection was performed 48 hours prior to luciferase detection (simultaneously with dsRNAs). To activate the Imd pathway, heat-killed E. coli (5×107) were added 4 hours prior to dual detection of the two luciferases.
Fly strains and infections
Flies were raised on standard culture medium at 25°C except if indicated. The insertion P[EPgy2]ash2EY03971 was obtained from BDSC. Transgenic lines were constructed in the P[UAST] vector [[23]]. The P[UAS-Usp2] transgene contains the CG14619-RA cDNA subcloned from SD02480 (DGRC). Heat-shock driven expression of Usp2 or Usp34 was achieved as described in [[17]]. For in vivo gene silencing, inverted repeats designed from the Heidelberg RNAi librarie (HFA, www.genomernai.org/) –that are similar as those used for S2 cell silencing- of Usp2 and Usp34 were cloned in the PWIZ vector [[49]]. Other silencing transgenes, P[UAS-Imd-IR] (#9253); P[UAS-Usp2-IR](#37930) and P[UAS-Usp34-IR] (#27517) were obtained from VDRC [[25]]. Importantly VDRC lines display different silencing sequences than our home-made constructs. For infection, fifty 3–5 days old males were pricked in the thorax with a thin needle that had been previously dipped in a concentrated overnight culture (OD600 #400) of E. coli or M. luteus (to measure activation of the Imd and Toll pathways respectively), or of K. pneumonia or E. faecalis, to measure flies survival.
Real-time quantitative PCR (RT-qPCR) analysis
Total RNAs were extracted from adult flies using Absolutely RNA Miniprep kit from Stratagene. For real-time PCR analysis, cDNAs were synthesized with AffinityScript QPCR cDNA Synthesis Kit (Stratagene). An amount of cDNA equivalent to 0.01 μg of total RNA was subjected to 40 cycles of PCR amplification consisting of a 10s incubation at 95°C and 30s at 60°C. Output was monitored using SYBR Green core reagents and the Mx3000P instrument (Stratagene). All the results were normalized to the rpl32 RNA level. The primer sequences were designed using PrimerQuest (http://eu.idtdna.com/Scitools/Applications/Primerquest/).
In vitro deubiquitinating assays
Mutation in the Usp2 coding sequence (resulting in the mutated protein USP2C540S) was introduced using QuickChange XL Site-directed Mutagenesis Kit (Stratagene). Wild type or mutated catalytic domains of USP2[AA 475 to 856] were coexpressed with Ub-β-gal fusion protein for 4 hours at 28°C in transformed E. coli XL1 Blue. Bacteria were lysed in 100 μL Laemmli solution. Samples were subjected to SDS-PAGE (6% gel) and western blotting with a rabbit anti β-Gal antibody (ROCKLAND).
Immunoprecipitation and immunoblotting
Co-immunoprecipitation were performed following standard procedures in S2 cotransfected cells with 10 μg of Myc tagged full length USP2 construct in pAc/HisB vector (Invitrogen) (USP2-Myc) and 10 μg of V5-tagged Imd full length or truncated constructs. Pull Down assays were performed in S2 cells transfected with full length Imd-V5 construct and lysed after 48 h. The lysate was employed in a GST-USP2 N-ter domain [AA 1 to 531] or C-ter domain [AA 475 to 856] pull down assays. Pull downs were blotted with antibody against V5 to detect bound Imd-V5.
Proteomic analysis (supporting data deposited in a database)
Protein digestion and nano-liquid chromatography (LC)–MS/MS analyses were performed as described in [[50]]. Only proteins identified specifically in the USP2 sample and not in the control one, with a minimum of 6 peptides and specific spectral counts above 6, were retained (indicated in green in Additional file 3: Table S2). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository [[51]] with the data set identifier PXD000881 and doi:10.6019/ PXD000881.
Competing interests
The author declare that they have no competing interests.
Authors’ contribution
EE, MM, and DT performed S2 cells screens, flies assays, RT-qPCR experiments and biochemical analyses. PV performed RT-qPCR experiments. YC did mass spectrometry analysis. EE, DT and ET help in the design of the experiments and in manuscript drafting. MOF designed the experiments, supervised the study and wrote the manuscript. All authors read and approved the final manuscript.
Additional files
Supplementary Material
Ubiquitin proteases screened on Imd and Toll pathways. List of Usps and corresponding CG numbers screened on Imd and Toll pathway in S2 cells with indication of human closest homolog gene(s). cDNA templates and primers used for dsRNA synthesis are indicated. All primers were designed from (http://www.genomernai.org/). Of note, the USP encoding gene CG8232 was not included in this study [[7],[19]]. The asterix (*) indicates the Usp genes not screened in [[7]]. The double asterix (**) indicates an alternative nomenclature used in [[7]].
Figure S1. Analysis of Usp36, Usp34 and Usp2 gene extinction by RT-qPCR in S2 cells treated for 48 h with the indicated dsRNAs dsGeneName). Figure S2. Activation of Drs promoter was monitored through the pDrs-luc reporter (normalised to pAct-luc) in transfected cells expressing pAc-Spz and treated for 48 h with the indicated dsRNAs. NI: no inducer. Figure S3. Analysis of Usp2 and Usp34 expression by RT-qPCR in HspGal4/+;UAS-Usp2/+ (Hsp>Usp2) or HspGal4/+; P{EPgy2}ash2EY03971 (Hsp>Usp34) compared to Hsp/+ flies at indicated time points after heat shock. Figure S4. Analysis of Usp2 (A), Usp34 (B) or Dpt (C,D) expression by RT-qPCR in total flies (A,C,D) or in dissected guts (B). Indicated silencing transgenes were induced by heat shock (HS-Gal4) (A) or in the gut (NP1-Gal4) (B) or in the fat body (c564-Gal4) (C,D). Figure S5.Dpt, AttA or DefC expression in c564-Gal4/Usp2-IR (#5 M) (A) or c564-Gal4/Usp34-IR (#1 M) (B,C) compared to c564-Gal4/+ flies. A,C: Flies were infected by E. coli by a septic injury (100% of activation fixed at 3 h post-infection). Figure S6. Analysis of Usp2 (A) or Usp34 (B) expression in flies by RT-qPCR at indicated time points following infection with E. coli or M. luteus. NI: not infected flies. Slight differences in gene expression were considered not biologically significant (below 1.6 fold). Figure S7. USP2 preferentially interacts with Imd-Nter. A. Representation of Imd full length (Imd-FL) and truncated constructs used in GST pull down assays. CD: catalytic domain in USP2, C540 catalytic cysteine, DD: death domain in Imd. B. S2 cells were transfected with pAc-USP2-Myc and lysed after 48 h. Cells lysates were pre-cleared and subjected to GST pull down using the indicated GST fusion proteins: Imd FL, Imd-Nter or the Imd-Cter. USP2-Myc was detected by western blot with anti-Myc antibodies.
List of USP2 interacting proteins identified by a Mass spectrometric analysis. Proteins identified specifically in the USP2 sample and not in the GST control one with a minimum of 6 peptides and specific spectral counts above 6 are indicated. The full mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository [[51]] with the data set identifier PXD000881 and doi:10.6019/ PXD000881.
Contributor Information
Elodie Engel, Email: elodie.engel@ujf-grenoble.fr.
Perrine Viargues, Email: perrine.viargues@gmail.com.
Magda Mortier, Email: magda.mortier@cea.fr.
Emmanuel Taillebourg, Email: emmanuel.taillebourg@cea.fr.
Yohann Couté, Email: yohann.coute@cea.fr.
Dominique Thevenon, Email: dominique.thevenon@cea.fr.
Marie-Odile Fauvarque, Email: mofauvarque@cea.fr.
Acknowledgments
We thank Dr Daniel Finley and Dr Gilles Courtois for powerful discussion, Dr Jean-Marc Reichhart for sending anti-Imd antibody, Dr Jean-Luc Imler for AttA-luciferase reporter construct. We are grateful to Nicole Assard, Dr Jacques Baudier, Dr Maxim Balakirev for scientific advices, Claire Bama and Christine Magnin for fly food and laboratory maintenance. The fly stocks were obtained from NIG-Fly Stock Center, Bloomington Drosophila Stock Center and VDRC. The cDNA clones were obtained from DGRC. This work was supported by the Fondation pour la Recherche contre le cancer (ARC n° SFI20111204045 to MOF), the CEA –including CEA-Irtelis doctoral grants (to EE and PV)- and the Agence Nationale pour la Recherche (ANR Investissement d’Avenir Infrastructures, ProFi project ANR-10-INBS-08-01 to YC).
References
- Ciechanover A. Tracing the history of the ubiquitin proteolytic system: the pioneering article. Biochem Biophys Res Commun. 2009;387:1–10. doi: 10.1016/j.bbrc.2009.06.065. [DOI] [PubMed] [Google Scholar]
- Clague MJ, Coulson JM, Urbe S. Cellular functions of the DUBs. J Cell Sci. 2012;125:277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S, Rajalingam K, Dikic I. Ubiquitylation in immune disorders and cancer: from molecular mechanisms to therapeutic implications. EMBO Mol Med. 2012;4:545–556. doi: 10.1002/emmm.201100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harhaj EW, Dixit VM. Regulation of NF-kappaB by deubiquitinases. Immunol Rev. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Tsou WL, Sheedlo MJ, Morrow ME, Blount JR, McGregor KM, Das C, Todi SV. Systematic analysis of the physiological importance of deubiquitinating enzymes. PLoS ONE. 2012;7:e43112. doi: 10.1371/journal.pone.0043112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chen ZJ. Regulation of NF-kappaB by ubiquitination. Curr Opin Immunol. 2013;25:4–12. doi: 10.1016/j.coi.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/S0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Swaminathan CP, Brown PH, Roychowdhury A, Wang Q, Guan R, Silverman N, Goldman WE, Boons GJ, Mariuzza RA. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs) Proc Natl Acad Sci U S A. 2006;103:684–689. doi: 10.1073/pnas.0507656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Meister M, Govind S, Georgel P, Steward R, Reichhart JM, Hoffmann JA. Functional analysis and regulation of nuclear import of dorsal during the immune response in Drosophila. EMBO J. 1995;14:536–545. doi: 10.1002/j.1460-2075.1995.tb07029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Reichhart JM, Hoffmann JA, Lemaitre B. In vivo regulation of the IkappaB homologue cactus during the immune response of Drosophila. J Biol Chem. 1998;273:10463–10469. doi: 10.1074/jbc.273.17.10463. [DOI] [PubMed] [Google Scholar]
- Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann JA, Bulet P. Differential display of peptides induced during the immune response of Drosophila: a matrix-assisted laser desorption ionization time-of-flight mass spectrometry study. Proc Natl Acad Sci U S A. 1998;95:11342–11347. doi: 10.1073/pnas.95.19.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci U S A. 2005;102:1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenon D, Engel E, Avet-Rochex A, Gottar M, Bergeret E, Tricoire H, Benaud C, Baudier J, Taillebourg E, Fauvarque MO. The Drosophila ubiquitin-specific protease dUSP36/Scny targets IMD to prevent constitutive immune signaling. Cell Host Microbe. 2009;6:309–320. doi: 10.1016/j.chom.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Li L, Anderson S, Secombe J, Eisenman RN. The Drosophila ubiquitin-specific protease Puffyeye regulates dMyc-mediated growth. Development. 2013;140:4776–4787. doi: 10.1242/dev.096941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu M, Su Y, Du J, Zhu AJ. A targeted in vivo RNAi screen reveals deubiquitinases as new regulators of Notch signaling. G3 (Bethesda) 2012;2:1563–1575. doi: 10.1534/g3.112.003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci U S A. 2000;97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D, Jung AC, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart J, Hoffmann JA. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. Embo J. 1998;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol Cell Biol. 2007;27:4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Hrdlicka L, Gibson M, Kiger A, Micchelli C, Schober M, Schock F, Perrimon N. Analysis of twenty-four Gal4 lines in Drosophila melanogaster. Genesis. 2002;34:51–57. doi: 10.1002/gene.10125. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. 2001;1:503–514. doi: 10.1016/S1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- Renatus M, Parrado SG, D’Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, Pierrat B, Riedl R, Vinzenz D, Worpenberg S, Kroemer M. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belote JM, Fortier E. Targeted expression of dominant negative proteasome mutants in Drosophila melanogaster. Genesis. 2002;34:80–82. doi: 10.1002/gene.10131. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Langmann C, Harden N, Aigaki T. The RING-finger scaffold protein Plenty of SH3s targets TAK1 to control immunity signalling in Drosophila. EMBO Rep. 2005;6:1082–1087. doi: 10.1038/sj.embor.7400537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Lim YM, Tsuda L, Nishida Y. Fat facets induces polyubiquitination of Imd and inhibits the innate immune response in Drosophila. Genes Cells. 2013;18:934–945. doi: 10.1111/gtc.12085. [DOI] [PubMed] [Google Scholar]
- Paquette N, Broemer M, Aggarwal K, Chen L, Husson M, Erturk-Hasdemir D, Reichhart JM, Meier P, Silverman N. Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol Cell. 2010;37:172–182. doi: 10.1016/j.molcel.2009.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Mao P, Smerdon MJ. Yeast deubiquitinase Ubp3 interacts with the 26 S proteasome to facilitate Rad4 degradation. J Biol Chem. 2010;285:37542–37550. doi: 10.1074/jbc.M110.170175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush RS, Cornwell WD, Uram JN, Lemaitre B. A ubiquitin-proteasome pathway represses the Drosophila immune deficiency signaling cascade. Curr Biol. 2002;12:1728–1737. doi: 10.1016/S0960-9822(02)01214-9. [DOI] [PubMed] [Google Scholar]
- Poalas K, Hatchi EM, Cordeiro N, Dubois SM, Leclair HM, Leveau C, Alexia C, Gavard J, Vazquez A, Bidere N. Negative regulation of NF-kappaB signaling in T lymphocytes by the ubiquitin-specific protease USP34. Cell Commun Signal. 2013;11:25. doi: 10.1186/1478-811X-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimerl F, Erhardt A, Sass G, Tiegs G. Down-regulation of the de-ubiquitinating enzyme ubiquitin-specific protease 2 contributes to tumor necrosis factor-alpha-induced hepatocyte survival. J Biol Chem. 2009;284:495–504. doi: 10.1074/jbc.M803533200. [DOI] [PubMed] [Google Scholar]
- Mahul-Mellier AL, Pazarentzos E, Datler C, Iwasawa R, AbuAli G, Lin B, Grimm S. De-ubiquitinating protease USP2a targets RIP1 and TRAF2 to mediate cell death by TNF. Cell Death Differ. 2012;19:891–899. doi: 10.1038/cdd.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzig M, Nickles D, Falschlehner C, Lehmann-Koch J, Straub BK, Roth W, Boutros M. An RNAi screen identifies USP2 as a factor required for TNF-alpha-induced NF-kappaB signaling. Int J Cancer. 2011;129:607–618. doi: 10.1002/ijc.26124. [DOI] [PubMed] [Google Scholar]
- Li Y, He X, Wang S, Shu HB, Liu Y. USP2a positively regulates TCR-induced NF-kappaB activation by bridging MALT1-TRAF6. Protein Cell. 2013;4:62–70. doi: 10.1007/s13238-012-2120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende-Vega N, Sparks A, Lane DP, Saville MK. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene. 2009;29:432–441. doi: 10.1038/onc.2009.330. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zanata SM, Kim J, Peterson MA, Di Vizio D, Chirieac LR, Pyne S, Agostini M, Freeman MR, Loda M. The ubiquitin-specific protease USP2a prevents endocytosis-mediated EGFR degradation. Oncogene. 2012;32:1660–1669. doi: 10.1038/onc.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WG, De Marzo AM, Yegnasubramanian S. USP2a activation of MYC in prostate cancer. Cancer Discov. 2012;2:206–207. doi: 10.1158/2159-8290.CD-12-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh KH, Yang SW, Park JM, Seol JH, Iemura S, Natsume T, Murata S, Tanaka K, Jeon YJ, Chung CH. Control of AIF-mediated cell death by antagonistic functions of CHIP ubiquitin E3 ligase and USP2 deubiquitinating enzyme. Cell Death Differ. 2011;18:1326–1336. doi: 10.1038/cdd.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Zhao W, Gu W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol Cell. 2009;36:469–476. doi: 10.1016/j.molcel.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Solomon LR, Pereda-Lopez A, Giranda VL, Luo Y, Johnson EF, Shoemaker AR, Leverson J, Liu X. Ubiquitin-specific cysteine protease 2a (USP2a) regulates the stability of Aurora-A. J Biol Chem. 2011;286:38960–38968. doi: 10.1074/jbc.M111.231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauszig-Delamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunol. 2002;3:91–97. doi: 10.1038/ni747. [DOI] [PubMed] [Google Scholar]
- Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/S1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Casabona MG, Vandenbrouck Y, Attree I, Coute Y. Proteomic characterization of Pseudomonas aeruginosa PAO1 inner membrane. Proteomics. 2013;13:2419–2423. doi: 10.1002/pmic.201200565. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O'Kelly G, Schoenegger A, Ovelleiro D, Perez-Riverol Y, Reisinger F, Rios D, Wang R, Hermjakob H. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ubiquitin proteases screened on Imd and Toll pathways. List of Usps and corresponding CG numbers screened on Imd and Toll pathway in S2 cells with indication of human closest homolog gene(s). cDNA templates and primers used for dsRNA synthesis are indicated. All primers were designed from (http://www.genomernai.org/). Of note, the USP encoding gene CG8232 was not included in this study [[7],[19]]. The asterix (*) indicates the Usp genes not screened in [[7]]. The double asterix (**) indicates an alternative nomenclature used in [[7]].
Figure S1. Analysis of Usp36, Usp34 and Usp2 gene extinction by RT-qPCR in S2 cells treated for 48 h with the indicated dsRNAs dsGeneName). Figure S2. Activation of Drs promoter was monitored through the pDrs-luc reporter (normalised to pAct-luc) in transfected cells expressing pAc-Spz and treated for 48 h with the indicated dsRNAs. NI: no inducer. Figure S3. Analysis of Usp2 and Usp34 expression by RT-qPCR in HspGal4/+;UAS-Usp2/+ (Hsp>Usp2) or HspGal4/+; P{EPgy2}ash2EY03971 (Hsp>Usp34) compared to Hsp/+ flies at indicated time points after heat shock. Figure S4. Analysis of Usp2 (A), Usp34 (B) or Dpt (C,D) expression by RT-qPCR in total flies (A,C,D) or in dissected guts (B). Indicated silencing transgenes were induced by heat shock (HS-Gal4) (A) or in the gut (NP1-Gal4) (B) or in the fat body (c564-Gal4) (C,D). Figure S5.Dpt, AttA or DefC expression in c564-Gal4/Usp2-IR (#5 M) (A) or c564-Gal4/Usp34-IR (#1 M) (B,C) compared to c564-Gal4/+ flies. A,C: Flies were infected by E. coli by a septic injury (100% of activation fixed at 3 h post-infection). Figure S6. Analysis of Usp2 (A) or Usp34 (B) expression in flies by RT-qPCR at indicated time points following infection with E. coli or M. luteus. NI: not infected flies. Slight differences in gene expression were considered not biologically significant (below 1.6 fold). Figure S7. USP2 preferentially interacts with Imd-Nter. A. Representation of Imd full length (Imd-FL) and truncated constructs used in GST pull down assays. CD: catalytic domain in USP2, C540 catalytic cysteine, DD: death domain in Imd. B. S2 cells were transfected with pAc-USP2-Myc and lysed after 48 h. Cells lysates were pre-cleared and subjected to GST pull down using the indicated GST fusion proteins: Imd FL, Imd-Nter or the Imd-Cter. USP2-Myc was detected by western blot with anti-Myc antibodies.
List of USP2 interacting proteins identified by a Mass spectrometric analysis. Proteins identified specifically in the USP2 sample and not in the GST control one with a minimum of 6 peptides and specific spectral counts above 6 are indicated. The full mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository [[51]] with the data set identifier PXD000881 and doi:10.6019/ PXD000881.


