FIGURE 1.
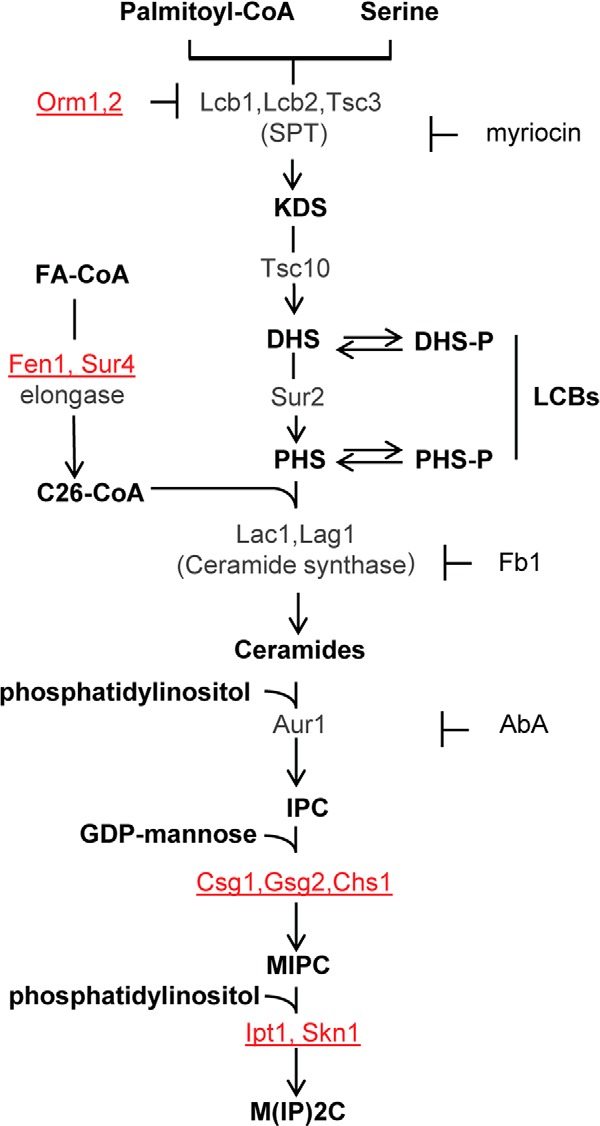
Schematic view of the sphingolipid metabolism in S. cerevisiae. Metabolic intermediates and complex sphingolipids are shown in bold; genes are indicated in gray or red. The genes indicated in red correspond to mutants resistant to DspA/E-mediated toxicity. Specific inhibitors of SPT (myriocin), ceramide synthase (Fb1), and IPC synthase (AbA) are also indicated. Sphingolipid biosynthesis starts in the endoplasmic reticulum with the condensation of serine and palmitoyl-CoA. This step is catalyzed by SPT to produce 3-ketodihydrosphingosine (KDS). Two related proteins, Lcb1 and Lcb2, heterodimerize to form the active SPT. A third protein, Tsc3, associates with the Lcb1/Lcb2 heterodimer and stimulates SPT activity several fold, whereas Orm1p and Orm2p proteins act as negative regulators of SPT. 3-Ketodihydrosphingosine is immediately reduced to DHS by an NADPH-dependent reaction catalyzed by an enzyme encoded by the TSC10 gene. DHS can then be hydroxylated by the Sur2 protein to give PHS. DHS and PHS are the yeast LCBs that are amide-linked to a very long chain fatty acid by the ceramide synthase to form ceramides. Ceramide synthase activity requires either of two redundant genes, LAC1 and LAG1. Ceramides are then modified in the Golgi apparatus to form complex sphingolipids. The first modification is the addition of myoinositol phosphate to form inositol-phosphoceramide (IPC). This reaction is catalyzed by IPC synthase whose activity requires the AUR1 gene. IPC is then mannosylated to yield mannose-inositol-phosphoceramide (MIPC), a reaction that requires the CSG1 and CSG2 genes. The terminal step in sphingolipid biosynthesis is the addition of inositol phosphate to MIPC to yield mannose-(inositol-P)2-phosphoceramide (M(IP)2C). The genes involved in this last reaction are IPT1 and SKN1. FA, fatty acyl.
