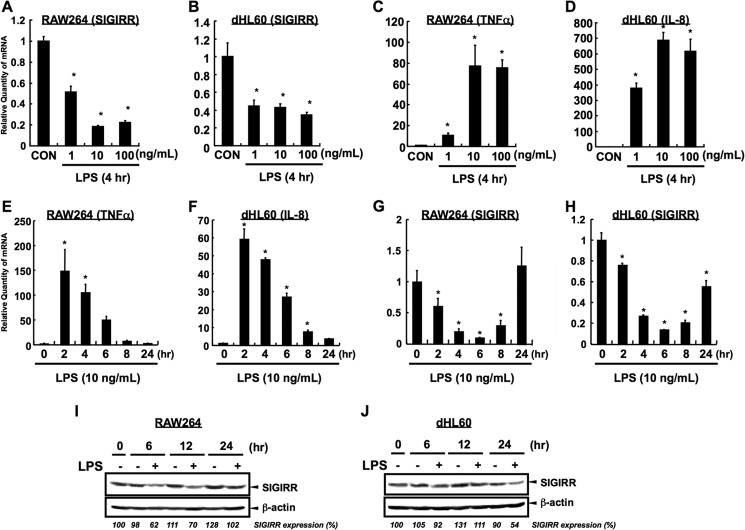
FIGURE 3.
LPS-dependent down-regulation in RAW264 and dHL60 cells. A–D, quantitative RT-PCR was carried out to detect the down-regulation of SIGIRR at mRNA level in RAW264 (A) and dHL60 (B) after LPS exposure in the indicated dose conditions. Quantitative RT-PCR was also carried out to detect the up-regulation of TNFα and IL-8 at mRNA level in RAW264 (C) and dHL60 (D), respectively. SIGIRR and cytokines mRNA levels were normalized to the 18 S rRNA mRNA levels as an internal control. *, p < 0.05, versus vehicle-treated cells; ANOVA with Dunnett's test (n = 3). E–H, time course analysis of cytokines (E and F) and SIGIRR (G and H) at mRNA level in RAW264 (E and G) and dHL60 (F and H) after LPS exposure in indicated time condition. SIGIRR and cytokines mRNA levels were normalized to the 18 S rRNA mRNA levels as an internal control. *, p < 0.05 versus vehicle-treated cells; ANOVA with Dunnett's test (n = 3). I and J, RAW264 (I) and dHL60 (J) cells were stimulated with 1 μg/ml LPS for indicated time periods, and the cell lysates were analyzed by Western blotting. Relative quantity of each band of SIGIRR protein was quantified and is shown as the % of band intensity of LPS non-treated group at 0 h.