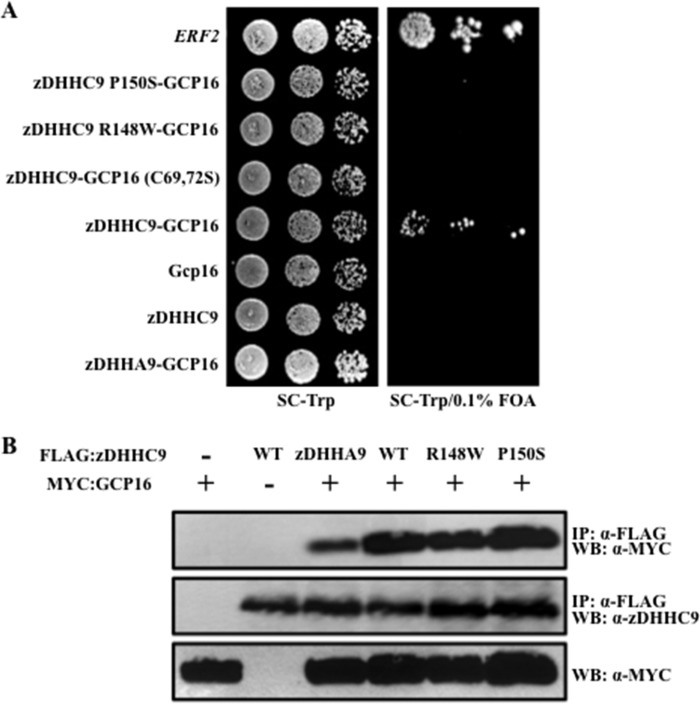
FIGURE 2.
zDHHC9-GCP16 interaction is required to suppress the loss of ERF2. A, to determine the ability of different zDHHC9 alleles and zDHHC9-GCP16 constructs to suppress the loss of ERF2 in S. cerevisiae, we performed a functional plasmid shuffle assay (22). Strain RJY1330 harboring plasmids expressing zDHHC9, zDHHC9-GCP16, zDHHC9 R148W-GCP16, zDHHC9 P150S-GCP16, zDHHC9-GCP16 (C69S,C72S), zDHHA9-GCP16, or ERF2 from the GAL1,10 promoter, were monitored for RAS2-independent growth on synthetic medium lacking tryptophan and supplemented with 5′-fluoroorotic acid (5′-FOA) to select for those strains capable of losing the URA3-linked RAS2 plasmid. Strains were initially grown in liquid synthetic medium lacking tryptophan and were spotted in 10-fold dilutions (50 × 103 initial cfu) on solid medium lacking tryptophan (top panel) and medium lacking tryptophan supplemented with 0.1% 5′-fluoroorotic acid (bottom panel). The plates were incubated for 4 days at 30 °C. B, mutations at zDHHC9 amino acid positions R148W and P150S do not interfere with the GCP16 interaction. Immunoprecipitates (IP) from whole cell extracts isolated from strain RJY1330 expressing zDHHC9-GCP16, zDHHC9, GCP16, zDHHA9-GCP16, zDHHC9 R148W-GCP16, and zDHHC9 P150S-GCP16 proteins were isolated using anti-FLAG-conjugated agarose. The immunoprecipitates were separated using SDS-PAGE (12%), transferred to nitrocellulose, and probed with antibodies to the c-Myc epitope (to identify myc-GCP16) or antibodies to the FLAG epitope (to identify zDHHC9). Western blot (WB) analysis of the whole cell extract utilizing antibodies to c-Myc was performed to demonstrate the presence of myc-GCP16 in the extracts.