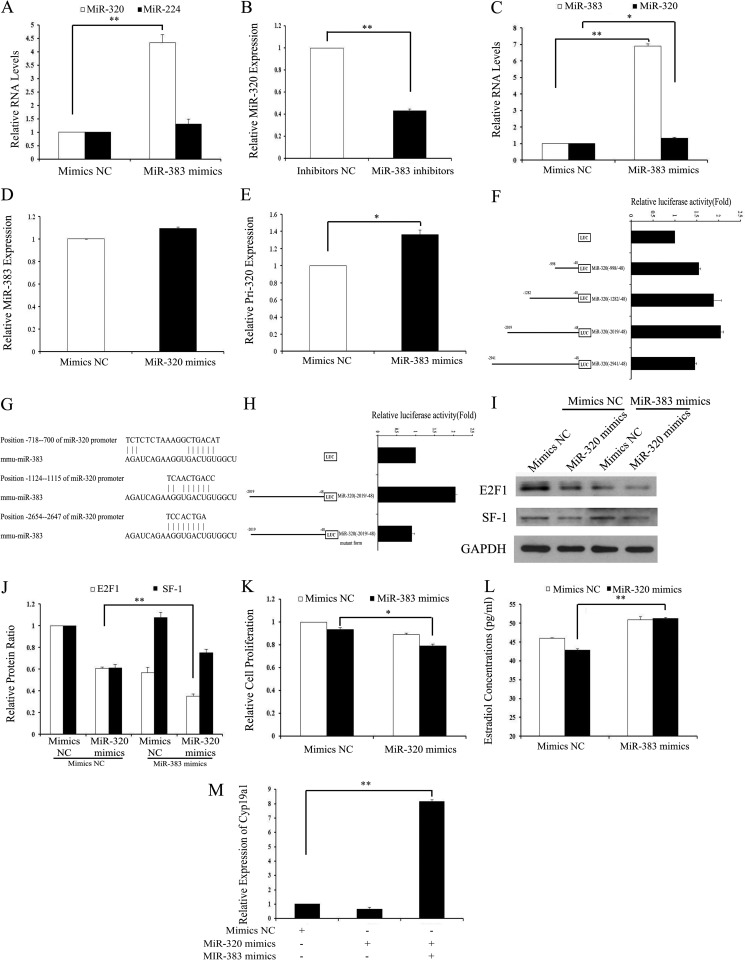
FIGURE 5.
miR-383 promotes the expression of miR-320 in GCs. A and B, miR-383 regulated the miR-320 levels. GCs were transfected with 120 nm miR-383 mimics/mimics NC (A) or 150 nm miR-383 inhibitors/inhibitors NC (B), and then the levels of miR-320 and miR-224 were determined after 48 h of transfection by real time PCR. C, miR-383 regulated the expression of miR-320 in the ovaries in vivo. Ovaries were injected with miR-383 mimics for 3 days as described above. Expression of miR-383 and miR-320 in the ovaries was determined by real time PCR. D, miR-320 had no effect on the expression of miR-383. GCs were transfected with 120 nm miR-320 mimics/mimics NC, and then the levels of miR-383 were determined after 48 h of transfection by real time PCR. E, miR-383 regulated the levels of primary miR-320. GCs were transfected with 120 nm miR-383 mimics/mimics NC, and then the level of primary miR-320 was determined after 48 h of transfection by real time PCR. F, identification of the minimal promoter region of miR-320. Left panel, schematic representation of 5′- or 3′-deleted fragments of miR-320 promoter in conjunction with the luciferase gene (LUC) in a pGL3-Basic vector. The nucleotides are numbered from the transcriptional starting site. Right panel, luciferase activities of the deleted miR-320 promoter constructs. HEK293T cells were co-transfected with miR-383 mimics and serial mouse miR-320 promoter-luciferase reporter deletion constructs. Relative promoter luciferase activity was calculated as a ratio of luciferase activity of the miR-320 promoter deletion constructs driven by miR-383 over the pGL3-Basic empty vector, which was arbitrarily set at 1. G, putative-binding sites for mmu-miR-383 are predicted in the 3000 bp of the miR-320 promoter. H, mutational analysis of miR-320 promoter-luciferase reporter constructs. miR-320 promoter regions (−2019/−48) containing mutations between −1124 and −1115 nucleotides were cloned into the luciferase reporter vector pGL3-Basic. I–K, overexpression of miR-383 enhanced miR-320-induced inhibition of E2F1 expression and proliferation suppression. The protein expression of E2F1/SF-1 (I and J) and cell viability (K) was analyzed after co-transfection with miR-320 mimics/mimics NC and miR-383 mimics/mimics NC into GCs. Representative Western blotting for E2F1 (upper bands), SF-1 (middle bands), and GAPDH (lower bands) (I) and the corresponding densitometric analysis (J) are shown. The ratio of the E2F1 band intensity over the GAPDH band intensity in controls was arbitrarily set at 1.0. L, overexpression of miR-383 rescued miR-320-induced E2 suppression. E2 were measured after co-transfection with miR-320 mimics/mimics NC and miR-383 mimics/mimics NC into GCs. M, overexpression of miR-383 rescued miR-320-induced suppression of CYP19A1 expression. The mRNA of Cyp19a1 was measured after co-transfection with miR-320 mimics/mimics NC and miR-383 mimics/mimics NC into GCs by real time PCR. A–F, H, and J–M, data represent the mean ± S.E. of three independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01.