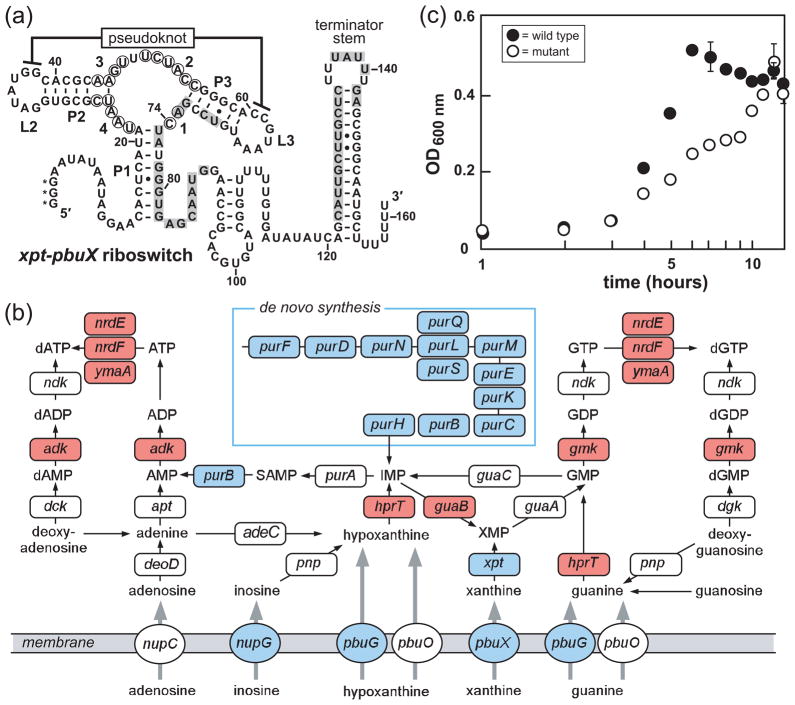
Figure 1.
Guanine riboswitches and guanine riboswitch-controlled genes in B. subtilis. a) Sequence and secondary structure of the xpt-pbuX guanine riboswitch from B. subtilis. Base-paired stems are denoted P1, P2 and P3, while loops are designated L2 and L3. Circled nucleotides identify those that undergo reduction of spontaneous cleavage during in-line probing reactions containing guanine concentrations above the KD, and positions are numbered as in Fig. 3b. Shaded nucleotides identify alternative pairing for the formation of the putative anti-terminator stem that may form when ligand is absent. Asterisks indicate nucleotides added to the RNA sequence to facilitate synthesis when this construct was prepared by in vitro transcription. b) Purine metabolic pathways in B. subtilis (47, 78, 79). Genes essential for B. subtilis growth (47) are highlighted in red, genes regulated by guanine riboswitches (37) are depicted in blue. c) The effects of the disruption of all four guanine riboswitch-controlled transcriptional units in B. subtilis. Growth curves of a wild-type B. subtilis strain (wild type) and a strain with all guanine riboswitch-controlled genes disrupted (mutant) are shown. The mutant strain carries knockouts of the genes controlled by the xpt-pbuX, pbuG, nupG riboswitches, and carries the pur operon downstream of a xylose-inducible promoter. The average of three independent growth assays is shown. Error bars depict the standard deviation of three separate assays, and were not added when the error was less than the width of the points.