Figure 3.
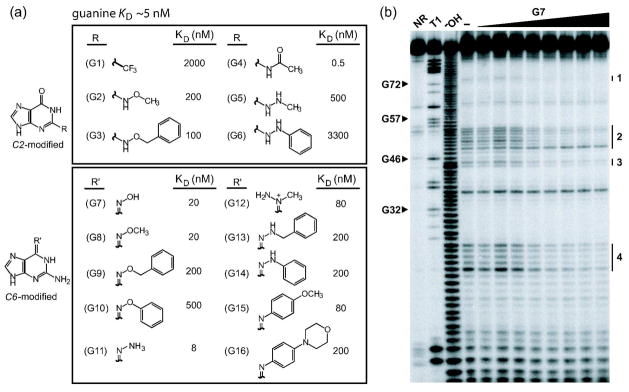
Guanine analog design and the binding characteristics of guanine analogs by the xpt-pbuX riboswitch in vitro. a) Guanine analog structures and KD values. The 16 guanine analogs are separated into two groups depending on whether chemical changes are made to the C2 or C6 position. b) PAGE analysis of the products of an in-line probing assay with the xpt aptamer (Figure 1, panel a). NR, T1, and −OH designate 5′ 32P-labeled RNAs subjected to no reaction, partial digest with RNase T1, or partial digestion with alkali, respectively. RNAs were incubated with G7 concentrations ranging from 0 to 1 μM, where (−) indicates incubation without ligand. Arrowheads identify several bands corresponding to nuclease cleavage after G residues (T1 lane) as designated according to the numbering system used in Figure 1, panel a. Regions 1 through 4 denote areas of structural modulation within the aptamer when ligand is bound.
