Fig. 1.
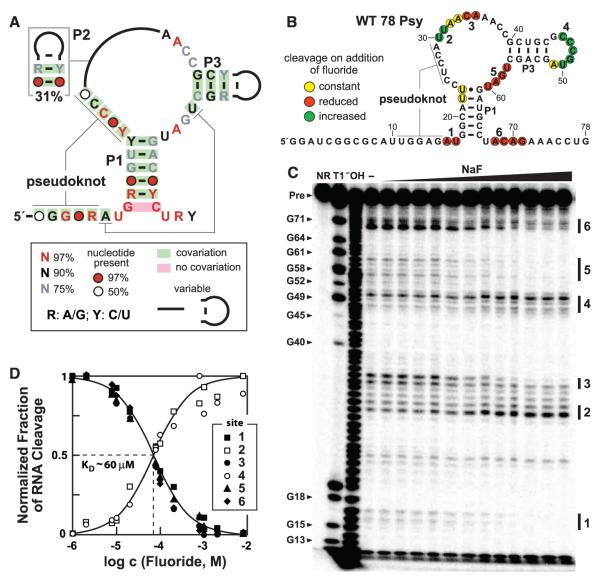
Fluoride binding by crcB motif RNAs. (A) Consensus sequence and structural model for crcB RNAs based on 2188 representatives from bacterial and archaeal species. P1, P2, P3 and pseudoknot labels identify extended base-paired substructures. (B) Sequence and secondary structure model for the WT 78 Psy RNA from P. syringae. Colored circles summarize the in-line probing results presented in (C). The two G residues preceding nucleotide 1 were added to facilitate RNA production. (C) Polyacrylamide gel electrophoresis analysis of an in-line probing assay with 78 Psy RNA and various amounts of fluoride. NR, T1, and −OH designate no reaction, partial digestion with RNase T1 (cleaves after guanosines), or partial digestion with hydroxide ions (cleaves after any nucleotide), respectively. Precursor RNA (Pre) band and some RNase T1 product bands are labeled (left). Locations of fluoride-mediated spontaneous RNA cleavage suppression (regions 1, 3, 5, 6) and enhancement (regions 2, 4) are identified by vertical bars. (D) Plot of the normalized fraction of RNA cleavage versus fluoride ion concentration from the data in (C). Curves represent those expected for one-to-one binding with a KD of 60 μM.