Figure 1. Riboswitch ligands and mechanisms.
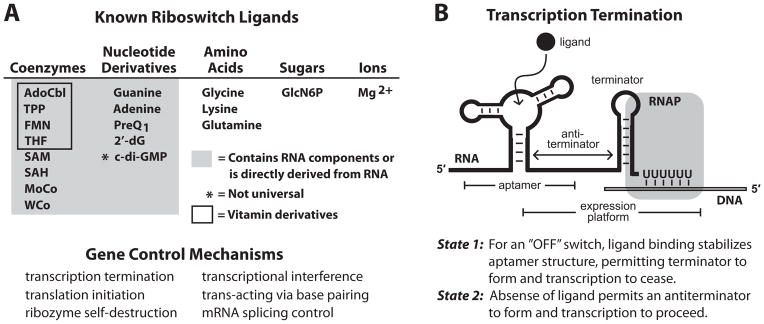
(A) (Top) List of the riboswitch ligands with biochemical and or genetic validation. (Bottom) Demonstrated mechanisms for riboswitch-mediated gene control.
(B) Schematic representation of the most common form of riboswitch-mediated gene regulation: transcription termination. Image depicts RNA polymerase (RNAP) in the act of transcribing the U-rich portion of an intrinsic transcription terminator stem in “State 1”, wherein ligand has been bound by the aptamer and transcription will terminate. Alternatively, in “State 2” (not shown), the absence of ligand allows nucleotides from the aptamer to form a competing anti-terminator stem that allows transcription to pass beyond the U-rich termination site. Other riboswitch mechanisms are described elsewhere (Breaker, 2011).
