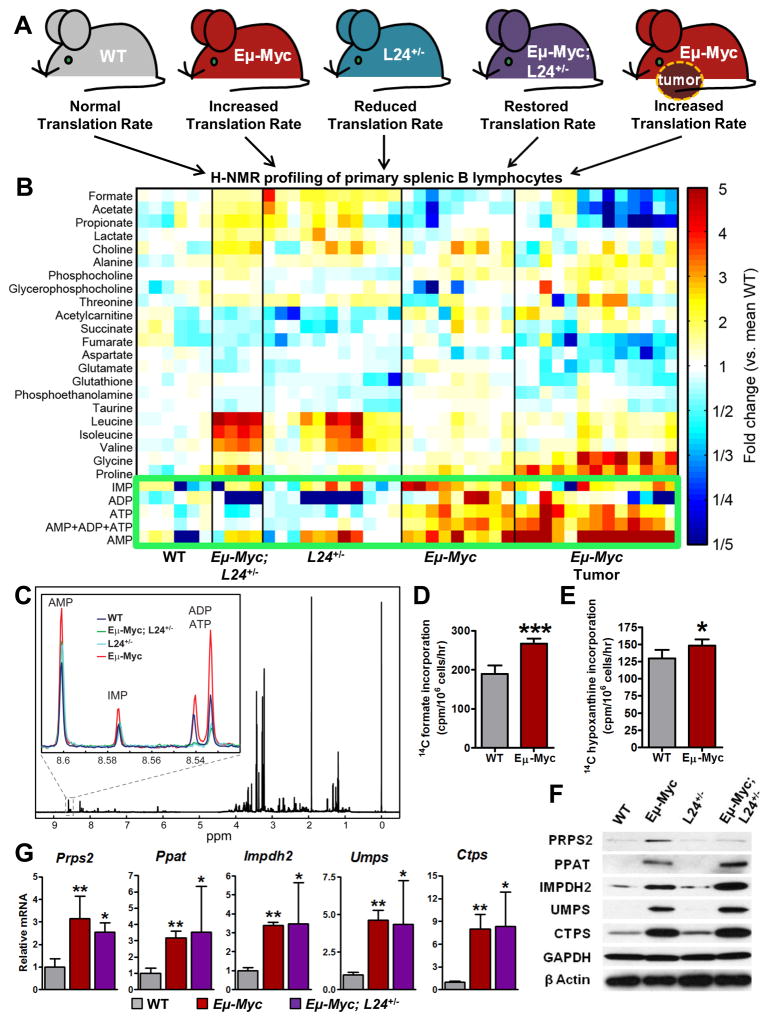
Figure 1. Unbiased Metabolomics Approach Reveals Myc-Dependent Translational Control of Purine Nucleotide Levels.
(A) Genetic approach to identify translationally regulated metabolites downstream of oncogenic Myc in B cells. (B) Intracellular steady state metabolite profiles of B cells isolated from wild-type (WT), Eμ-Myc/+ premalignancy, Rpl24BST/+, (L24+/-)Eμ-Myc/+;Rpl24BST/+, and Eμ-Myc/+ tumor-bearing mice. (C) Representative 1H NMR spectra of extracted metabolites from B cells with (inset) zoomed in view of region between 8.52–8.61ppm containing representative purine profiles from indicated genotypes. (D) Metabolic flux through de novo purine synthesis pathway measured by [14C] formate incorporation in WT and Eμ-Myc/+derived B cells. (E) Metabolic flux through purine salvage pathway measured by [8-14C] hypoxanthine incorporation in WT and Eμ-Myc/+-derived B cells. (F) Western blot analysis of indicated nucleotide biosynthesis enzymes from B cells derived from 5 week old WT, Eμ-Myc/+, Rpl24BST/+, and Eμ-Myc/+;Rpl24BST/+ mice. (G) qRT-PCR analysis of mRNA levels of indicated enzymes from mice as in (F). Error bars represent standard deviation, N=6 for (D) and (E), N=4 for (G)***P<0.001, **P<0.01, *P<0.05 by student’s t-test. See also Figure S1.