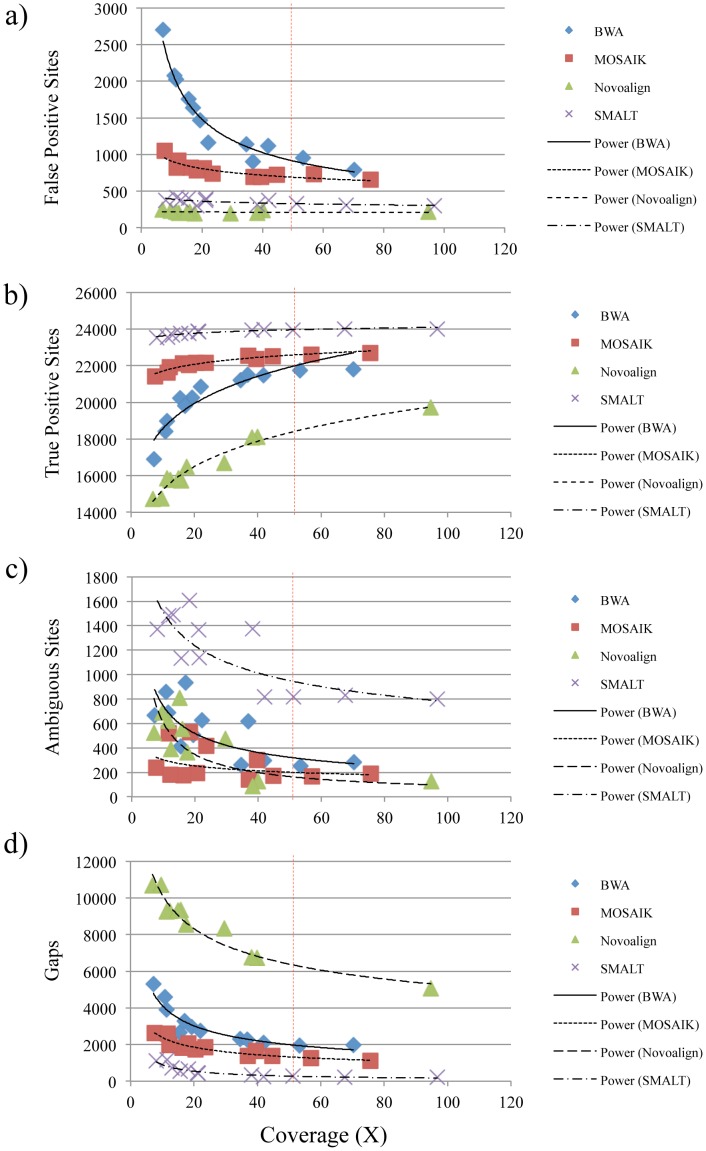
Figure 3. Comparison of consensus sequences calculated from alignments of Illumina MiSeq short-read data to a non-identical reference with four reference-guided assemblers.
Genomic DNA from the Listeriosis Reference Service for Canada's (LRS) Listeria monocytogenes strain 08-5578 subculture was sequenced and indexed twelve times. The resulting reads were aligned with four reference-guided assemblers (BWA, MOSAIK, Novoalign, and SMALT) using an L. monocytogenes strain EGD-e chromosome sequence obtained from the National Center for Biotechnology Information (NCBI) archive as a reference. The NCBI strain EGD-e chromosome sequence differs from the LRS strain 08-5578 chromosome sequence at 25,347 nucleotide positions. The numbers of false positive sites (a), true positive sites (b), ambiguous sites (c), and gaps (d) present in the resulting consensus sequences relative to the calculated coverage of each assembly are shown. Lines were calculated with power regression and smoothed using a Catmull-Rom Spline.