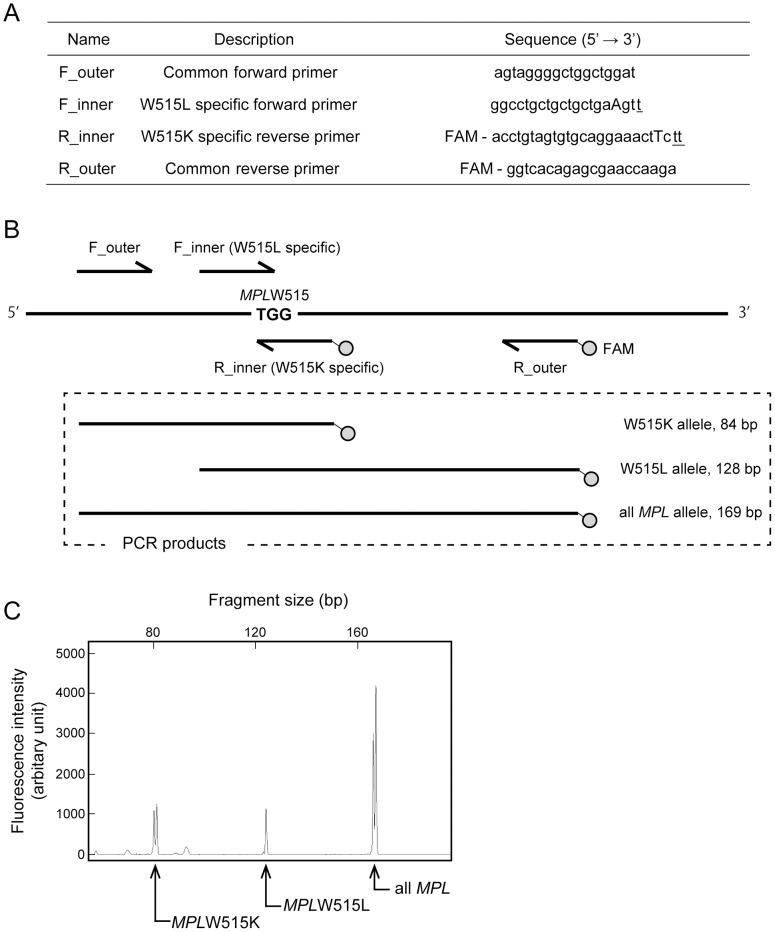
Figure 1. Detection of MPLW515L/K mutations using DARMS-PCR.
(A) The primers used in the DARMS-PCR assay. The two inner primers harbored sequences (underlined) that matched MPLW515L or W515K, but not the wild-type allele. Other mismatches (capital letters) were introduced into the inner primers to reduce the annealing of the mutant-specific primers to the wild-type sequence. The reverse primers were labeled with FAM (5-carboxyfluorescein hydrate) at the 5′ terminus. (B) A schematic representation of DARMS-PCR products. The two outer primers were designed to generate a 169-base-pair (bp) PCR product from all MPL alleles. The F_inner and R_inner primers annealed specifically to the MPLW515L and W515K alleles, respectively; in combination with the outer primers, they generated 84- and 128-bp PCR products, respectively. From a mutant allele, both 169-bp and 84- or 128-bp fragments were amplified, while, only the 169-bp fragment was generated from the wild-type allele. (C) Demonstration of DARMS-PCR. A capillary electropherogram of DARMS-PCR products showing three peaks derived from wild-type MPL, W515L, and W515K. This result was obtained when PCR was performed with a standard DNA mixture containing equal ratios of MPL wild-type, W515L, and W515K alleles with a total copy number of 105. The horizontal axis represents the fragment length, and the vertical axis represents the fluorescence intensity.