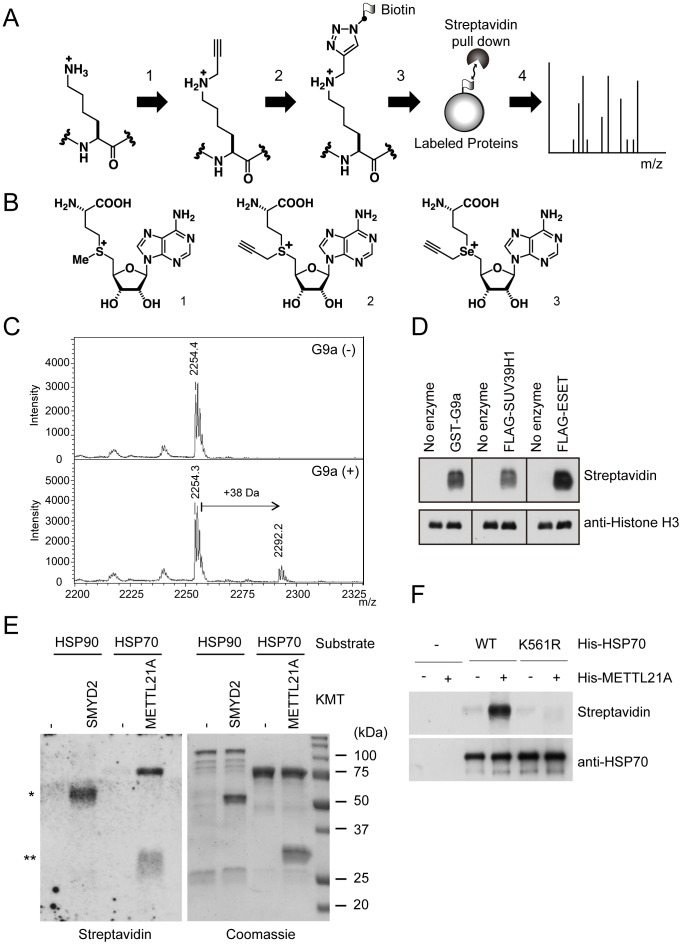
Figure 1. ProSeAM, a synthetic SAM analog, has a wide spectrum of reactivity for histones and non-histone substrates.
A, Schematic overview for analyzing lysine methylation. A synthetic cofactor was used to transfer an alkyne moiety to the ε-amino group of lysine by KMTs (1). The modified proteins were tagged with biotin via CuAAC reaction (2). Tagged-proteins in the crude lysates were pulled down with affinity beads (3), and the precipitants were further analyzed with a LC-MS apparatus (4). B, Chemical structure of SAM (1), propargylated SAM (2) and ProSeAM (3). C, H3 peptide (1-21 a.a.) and ProSeAM was incubated with or without GST-G9a at 20°C for 2 h, then the peptide was analyzed by MALDI-TOF MS. D, full-length Histone H3 (1 µg) and ProSeAM (500 µM) were incubated with indicated KMTs (0.5 µg) for 2 h at 20°C. The histones were separated by SDS-PAGE, transferred to a nitrocellulose membrane and probed with streptavidin-HRP (top) or anti-Histone H3 antibody (bottom). E, The non-histone substrates His-HSP90 and His-HSP70 (1 µg) were incubated with His-SMYD2 and His-METTL21A (1 µg), respectively. After the reaction, proteins were separated by SDS-PAGE (right). Their modifications were detected by western blotting with streptavidin-HRP as in Fig. 1D. *and ** showed automodification of SMYD2 and METTL21A, respectively (left). F, His-HSP70 (WT and K561R) were incubated with or without His-METTL21A in the presence of ProSeAM for 2 h at 20°C. Modified proteins were biotinylated and detected with streptavidin-HRP (top) or anti-HSP70 antibody for the loading control (bottom).