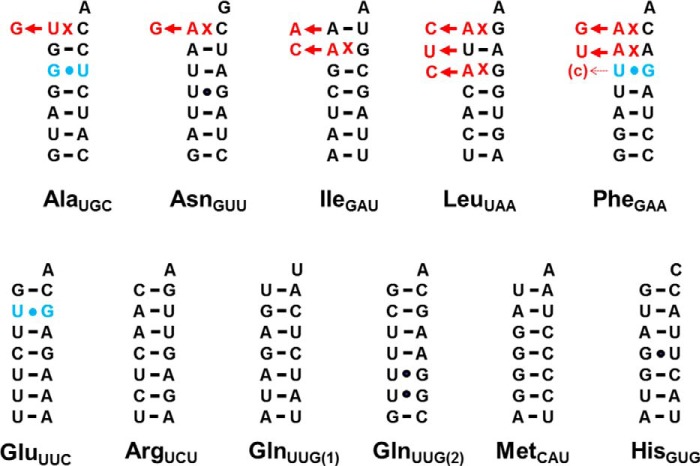
FIGURE 3.
Summary of verified mt-tRNA editing events in P. pallidum. Aminoacyl-acceptor stem sequences of 11 mtDNA-encoded tRNA species in P. pallidum are shown. Clones containing the corresponding mt-tRNA genes were isolated from P. pallidum strain PPHU8 mtDNA and sequenced (36). Sequence comparison indicates that the mitochondrial genome of strain PPHU8 is essentially identical to that of P. pallidum strain PN500 (GenBank accession number EU275726.1). The top row includes mt-tRNAs predicted to be substrates for 5′-editing (based on the presence of identifiable mismatches) prior to this work; the bottom row includes mt-tRNAs that do not contain mismatched nucleotides in the first three positions of their aminoacyl-acceptor stem. Mismatched nucleotides in the originally encoded mt-tRNA genes are indicated in red; alterations that result in Watson-Crick base-paired nucleotides verified here are indicated by red nucleotides shown to the left of each arrow. U-G/G-U base pairs in any of the first three positions of the aminoacyl-acceptor stem are indicated in blue; changes at any of these nucleotides, if observed here, are also indicated by red nucleotides shown to the left of each arrow. Partial editing is indicated by small arrows with the replacement nucleotide that is observed in some but not all clones indicated in parentheses next to the arrowhead.